Abstract
The message is loud and clear. MicroRNA-96, one in a cluster of three related neurosensory microRNAs, is crucial to the development and maintenance of inner ear hair cells and hearing in mice and humans. Two recent studies show that mutations in the critical seed region of the microRNA underlie the cause of hair cell degeneration and progressive hearing loss. Other recent reports reveal the general requirement of microRNAs for sensory epithelial development and maintenance in Dicer knockout mouse ear. The challenge begins to determine whether microRNAs will resonate as therapeutic agents or target molecules to preserve or restore hearing.
Something to hear about
MicroRNAs (miRNAs) have made quite a clatter since their widespread eukaryotic distribution, evolutionary conservation, and biological significance have become apparent in recent years [1, 2]. These small RNAs function to post-transcriptionally repress target messenger RNA (mRNA) expression, often producing subtle changes in protein expression [3, 4]. Nevertheless, the net effect levied by individual or combined miRNA function contributes substantially to normal processes in development and aberrant processes in disease. Such is the case in cancer biology [5], where changes in miRNA expression or mutations affecting miRNA function underlie biological mechanisms supporting cell growth, suggesting that miRNAs might serve as efficacious agents or target molecules in treating disease.
Chances are that you have heard the clatter, the literal fact of which can be attributed in part to miRNA function in mechanosensory hair cells of the inner ear. Two recent studies published in Nature Genetics show that mutations in miRNA-96 (miR-96) are responsible for non-syndromic progressive hearing loss in humans and mice [6, 7]. These studies importantly establish the first known Mendelian disease associated with miRNA function, and place a miRNA gene among the ranks of more than 100 other genetic loci associated with heritable deafness or hearing impairment [8], reflecting the anatomical and functional complexity of the inner ear.
miR-96 is one in a cluster of three related miRNAs in vertebrate organisms that additionally includes miR-182 and miR-183 (miR-96/182/183) [9]. These conserved miRNAs are processed from a common primary transcript [10–12] and are thus co-expressed in neurosensory cells and organs, including photoreceptor cells in the eye and hair cells in the ear [10, 11, 13]. Moreover, they are homologous in sequence to one another and to other miRNAs (miR-228 and miR-263b) exhibiting neurosensory expression in invertebrate organisms [14]. The strong evolutionary conservation and neurosensory expression of the broader family suggest that these miRNAs play critical roles in the development and function of sensory cells. Nevertheless, each miRNA possesses a distinct sequence difference in its seed region, the portion of the miRNA (nucleotides 2 to 8) that interacts with target mRNAs [15], implying the miRNAs serve non-overlapping functions that are further expected to depend upon target gene expression in specific cell types.
Silencing effect of one miRNA
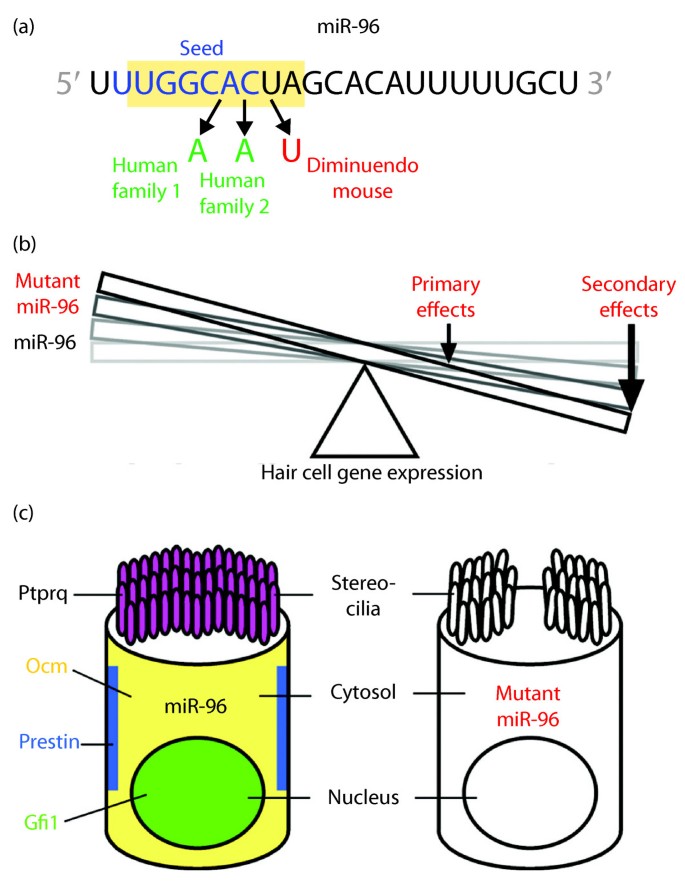
The significance of miR-96 expression and function in mechanosensory hair cells of the inner ear is revealed and fortified by both the similarity and distinction of results from genetic studies of mouse and human progressive hearing loss. Lewis et al. [7] report their findings on the diminuendo (Dmdo) mouse, a semidominant mutant allele identified in a screen for genes that cause deafness. The allele mapped to a region of chromosome 6 that includes the miR-96 gene (Mirn96), in which an A-to-T transversion affects the corresponding mutation at position 7 of the mature miRNA within the seed sequence (Figure 1a). Heterozygous Dmdo mutants exhibit a progressive and substantial hearing loss that correlates with stereocilia degeneration and auditory hair cell loss in the cochlea, whereas homozygous Dmdo mutants exhibit earlier degeneration leading to complete hair cell loss and profound deafness. In situ hybridization detecting the mature miRNA demonstrates that the mutation does not affect processing, suggesting that mutant miR-96 might cause hearing loss through a concomitant loss of function and gain of function.
MicroRNA-96 mutations effect changes in hair cell gene expression, hair cell degeneration and progressive hearing loss. (a) MicroRNA-96 sequence and mutations within the seed region (blue) found in humans (green) and the diminuendo mouse (red). (b) The balance of hair cell gene expression is offset by the diminuendo mutation in miR-96, where primary effects on target gene expression appear less in magnitude than secondary effects on the expression of other genes crucial to hair cell function. (c) Loss of crucial hair cell protein expression in diminuendo mouse contributes to hair cell degeneration and ultimate demise. The normal subcellular localization of hair cell proteins absent in diminuendo mouse is indicated.
Congruent with the mouse study, Mencía et al. [6] report their further analysis of a human autosomal dominant deafness locus (DFNA50) that mapped to a region of chromosome 7 that includes the miR-96 gene (MIR96) [16]. Analysis of the original DFNA50 family and hundreds of other genetically undiagnosed hearing-impaired families and normal hearing controls established two families with distinct mutations in miR-96 that segregate with the hearing loss phenotype [6]. Each affected family member possesses either a G-to-A transition or C-to-A transversion that respectively affects the corresponding mutation at position 5 or 6 of the mature miRNA within the seed sequence (Figure 1a). In vitro analysis demonstrates that the mutations affect processing and substantially diminish production of mature miRNA, suggesting that the hearing loss in humans is caused largely by miRNA loss of function. Overall, the commonality of hearing loss effected by different mouse and human mutations in the seed region of miR-96 [6, 7] strongly suggests the phenotype is caused by miRNA loss of function rather than gain of function per se. Moreover, the effect of miR-96 haploinsufficiency suggests that hair cells are extremely sensitive to perturbation of miRNA levels and that miR-96 is a dynamic regulator of hair cell gene expression.
Both studies [6, 7] examined the influence of miR-96 on a common set of predicted target genes expressed in the inner ear, including Aqp5 (an aquaporin), Celsr2 (a cadherin), Myrip (a myosin-interacting protein), Odf2 (a component of cilia), and Ryk (an orphan receptor associated with ephrin receptors). In vitro analysis demonstrates that miR-96 effects a mild but significant reduction in expression of predicted target-reporter gene constructs that is not reproduced by mutant miR-96 [6, 7]. However, immunohistochemical analysis in diminuendo mouse provided no evidence for changes in protein levels [7]. The results are consistent with proteomic and transcriptomic studies showing that miRNAs effect many mild changes in target gene expression [3, 4] that might not be detectible by less sensitive methodologies. Nevertheless, microarray analysis of gene expression in normal and diminuendo mouse cochlear sensory epithelium (that is, organ of Corti) demonstrates that changes in transcript levels correlate with miR-96 loss of function and mutant miR-96 gain of function [7]. Assessment using the Sylamer algorithm [17] shows that transcripts upregulated in diminuendo mouse are relatively enriched for complementary seed matches to miR-96, consistent with loss of function, whereas downregulated transcripts are relatively enriched for complementary seed matches to mutant miR-96, consistent with gain of function [7].
More striking than the subtle changes in predicted target gene expression (primary effects) are changes in the expression of other genes that do not possess complementary seed matches to normal or mutant miR-96 (secondary effects; Figure 1b). Immunohistochemical analysis of diminuendo mouse hair cells [7] shows drastic reduction in expression of genes crucial to hair cell function and survival, including Ptprq (required for stereocilia maturation), Ocm (calcium ion binding), Prestin (hair cell motor protein), and Gfi1 (hair cell transcription factor) (Figure 1c). Such results are not general features of degenerating hair cells in other mutant mouse models [7], suggesting that miR-96 specifically regulates a complex network of pathways supporting hair cell development, maturation and survival.
Considering the evolutionary conservation and expression of miR-96/182/183 in neurosensory hair cells and photo-receptors, it is noteworthy that individuals with hearing loss affected by mutations in miR-96 show no deficit in ophthalmologic function [6]. This suggests that each miRNA could fulfill a substantially different role in different neuro-sensory cells to varying degrees of functional significance. Interestingly, targeted deletion of miR-182 was recently reported to have no effect on the histological development of the retina in young mice [18]. However, the effect that miR-182 knockout has on the inner ear, or its possible functional deficits, remain to be elucidated. Consequently, there is much to be done to assess whether each miRNA in the miR-96/182/183 cluster might have a differential effect in the development and function of neurosensory cells in different organs.
Goodbye miRNAs, goodbye ear
The importance of miRNAs to hair cell development and maintenance is further corroborated by two recent studies [19, 20] that examined the effect of tissue-specific Dicer conditional knockout (CKO), which ablates mature miRNA production [21]. Developmentally early Dicer CKO and depletion of miRNAs throughout the inner ear results in severe morphohistological defects in sensory epithelia and hair cells, where the extent of hair cell development is shown to correlate with residual miR-183 expression [19]. Relatively later hair cell and sensory epithelium-specific Dicer CKO results in initial formation but rapid degeneration of hair cells, and deafness [20]. Interestingly, the morphology of miRNA-depleted hair cells in each model shows a high degree of similarity [19, 20]. Results from these studies lend further credence to the significance of miR-96/182/183 among other hair cell miRNAs [20] in hair cell development, maintenance and function [22].
Micromanaging hair cell survival or regeneration
The apparent biological potency of certain miRNAs such as miR-96 poses something of a double-edged situation with regard to their practical utility as therapeutic agents or targets. The delivery or inhibition [23] of miRNAs could prove efficacious or deleterious, depending on the pharmacokinetic and pharmacodynamic properties of the administration. One might anticipate that the use of any such agents in situ will require methodologies for cell-specific delivery and regulated dose and/or function to avoid perturbing the natural balance of other miRNA-mediated effects on the transcriptome. Nevertheless, understanding how miRNAs manage the business of gene expression in normal or degenerating hair cells is certain to provide routes of discovery to novel target molecules or biological processes that might otherwise be manipulated using traditional small-molecule pharmaceuticals to influence hair cell maintenance and survival. Moreover, appreciating how miRNAs affect normal differentiation and maturation of functional hair cells might contribute to in vivo or in vitro methodologies for, respectively, coaxing supporting cells or induced pluripotent stem (iPS) cells to undergo hair cell differentiation [24]. Regardless of strategy, any utilization of miRNAs to effect hair cell survival or regeneration is likely to be a long story, but one that just might be worth hearing.
Abbreviations
- CKO:
-
conditional knockout
- iPS:
-
induced pluripotent stem (cell)
- mRNA:
-
messenger RNA
- miRNA or miR:
-
microRNA.
References
Kloosterman WP, Plasterk RH: The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006, 11: 441-450. 10.1016/j.devcel.2006.09.009
Makeyev EV, Maniatis T: Multilevel regulation of gene expression by microRNAs. Science. 2008, 319: 1789-1790. 10.1126/science.1152326
Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N: Widespread changes in protein synthesis induced by microRNAs. Nature. 2008, 455: 58-63. 10.1038/nature07228
Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP: The impact of microRNAs on protein output. Nature. 2008, 455: 64-71. 10.1038/nature07242
Blenkiron C, Miska EA: miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum Mol Genet. 2007, 16: R106-R113. 10.1093/hmg/ddm056
Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, Del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA: Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009, 41: 609-613. 10.1038/ng.355
Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP: An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009, 41: 614-618. 10.1038/ng.369
Nance WE: The genetics of deafness. Ment Retard Dev Disabil Res Rev. 2003, 9: 109-119. 10.1002/mrdd.10067
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ: miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008, 36: D154-158. 10.1093/nar/gkm952
Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA: MicroRNA gene expression in the mouse inner ear. Brain Res. 2006, 1111: 95-104. 10.1016/j.brainres.2006.07.006
Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D: MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007, 282: 25053-25066. 10.1074/jbc.M700501200
Saini HK, Enright AJ, Griffiths-Jones S: Annotation of mammalian primary microRNAs. BMC Genomics. 2008, 9: 564- 10.1186/1471-2164-9-323
Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH: MicroRNA expression in zebrafish embryonic development. Science. 2005, 309: 310-311. 10.1126/science.1114519
Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA: MicroRNA-183 family conservation and expression in ciliated neurosensory organs. Evol Dev. 2008, 10: 106-113.
Lewis BP, Burge CB, Bartel DP: Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005, 120: 15-20. 10.1016/j.cell.2004.12.035
Modamio-Høybjør S, Moreno-Pelayo MA, Mencía A, del Castillo I, Chardenoux S, Morais D, Lathrop M, Petit C, Moreno F: A novel locus for autosomal dominant nonsyndromic hearing loss, DFNA50, maps to chromosome 7q32 between the DFNB17 and DFNB13 deafness loci. J Med Genet. 2004, 41: e14- 10.1136/jmg.2003.012500
van Dongen S, Abreu-Goodger C, Enright AJ: Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods. 2008, 5: 1023-1025. 10.1038/nmeth.1267
Jin ZB, Hirokawa G, Gui L, Takahashi R, Osakada F, Hiura Y, Takahashi M, Yasuhara O, Iwai N: Targeted deletion of miR-182, an abundant retinal microRNA. Mol Vis. 2009, 15: 523-533.
Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD: Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009, 328: 328-341. 10.1016/j.ydbio.2009.01.037
Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, Avraham KB: MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci USA. 2009, 106: 7915-7920. 10.1073/pnas.0808468106
Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ: The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005, 102: 10898-10903. 10.1073/pnas.0504834102
Soukup GA: Little but loud: small RNAs have a resounding affect on ear development. Brain Res. 2009, doi:10.1016/j.brainres.2009.02.027.
Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M: Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005, 438: 685-689. 10.1038/nature04303
Beisel K, Hansen L, Soukup G, Fritzsch B: Regenerating cochlear hair cells: quo vadis stem cell. Cell Tissue Res. 2008, 333: 373-379. 10.1007/s00441-008-0639-z
Acknowledgements
The authors' research in the area has been funded by grants from the National Institutes of Health, National Center for Research Resources, Centers of Biomedical Research Excellence (grant number P20RR018788); National Institute on Deafness and Other Communication Disorders, Ruth L Kirschstein National Research Service Award (grant number F32DC008253); and Nebraska State LB692.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Both authors contributed to the conceptualization and writing of the manuscript and have approved the final version.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Weston, M.D., Soukup, G.A. MicroRNAs sound off. Genome Med 1, 59 (2009). https://doi.org/10.1186/gm59
Published:
DOI: https://doi.org/10.1186/gm59