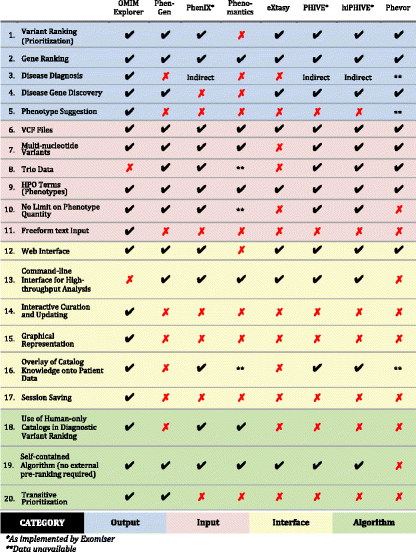
- 1. Ranking of input variants. 2. Ranking of genes containing input variants. 3. Ranking of diseases. 4. Identification of gene candidates for causal association with input phenotypes. 5. Identification of phenotypes that may help clarify or distinguish among top rankings. 6. Acceptance of variant sets as VCF (variant call format) files. 7. Inclusion of multi-nucleotide (insertion/deletion/frameshift) variants in computational prioritization. 8. Support for integration of family VCFs to distinguish between transmitted and de novo variation. 9. Acceptance of phenotypic query descriptors as HPO (Human Phenotype Ontology) terms. 10. Absence of limit on quantity of input phenotypes (HPO terms) supplied. 11. Acceptance of unstructured text from which input phenotypes are computationally extracted. 12. Accessibility via a web browser. 13. Accessibility via a command line API (application programming interface), which facilitates automated batch submission of distinct case queries. 14. Immediate update of outputs in response to changes in input or analysis configuration, including diagnostic exclusion, without repeating the entire input and analysis process. 15. Pictorial representation of output, in addition to tabular representation. 16. Graphical or tabular juxtaposition of outputs with input-specific catalog data (input variants hosted by gene, causal links between diseases and gene, phenotypes annotated to disease, known modes of inheritance of disease, etc.). 17. Export of input and configuration data in a file that can be subsequently imported and modified, and from which result outputs can be regenerated. 18. Restriction to use of only human data catalogs (known direct and ontological associations between diseases, genes, and phenotypes) in differential disease diagnosis and variant rank estimation for clinical decision support. 19. Calculation of disease and variant rankings without the use of externally-computed phenotype-based rankings. 20. Deductive-reasoning-based variant ranking through inference of host gene phenotypic relevance from semantic similarities of intact diseases to which genes are causally linked
