Fig. 1
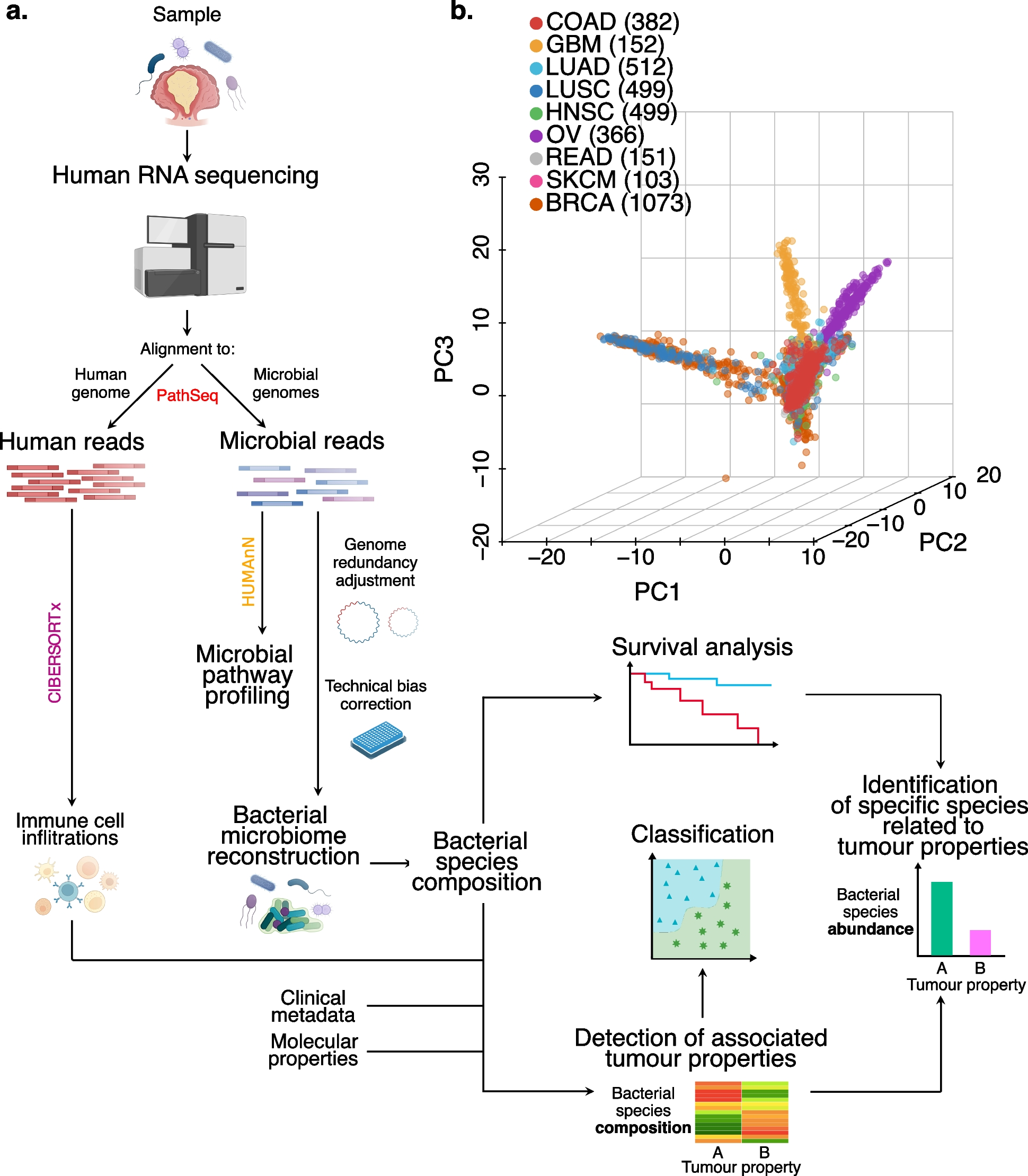
General overview on bacteria reconstructed microbiomes. a Summary of the microbial reconstruction workflow to detect clinical and molecular associations with bacteria. Each RNA sequencing BAM file was analysed by this workflow: after removing the human sequences, the rest of the reads were aligned to a set of microbial genomes from the National Center for Biotechnology Information (NCBI) and assigned to one or more species. The last step to reconstruct the microbiome of the samples involves a batch effect detection that identifies the influence of available technical properties on the reconstructed microbiome composition. Microbiome profiles are then corrected accordingly for the most relevant batch effects. Finally, the corrected microbiome profiles were tested for associations with clinical and molecular properties of the tumour, survival analysis and property classification. For the properties associated with microbiome composition, the bacterial composition underwent a property classification approach, while specific bacterial species were detected as linked to the property levels and the prognosis. Colon adenocarcinoma (COAD) bacterial reads were pooled into left and right-sided, CMS1 and CMSs pooled and mutation burden high and low and underwent pathway profiling to compare metabolic differences between the sides of the colon, CMSs and mutational burden levels, respectively. b Principal component analysis on all the reconstructed bacterial microbiomes of the cancer types analysed. GBM, glioblastoma multiforme; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; HNSC, head and neck squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; READ, rectum adenocarcinoma; SKCM, skin cutaneous melanoma; BRCA, breast invasive carcinoma. Number of samples analysed in brackets
