- Research
- Open access
- Published:
Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores
Genome Medicine volume 11, Article number: 74 (2019)
Abstract
Background
Inherited susceptibility to common, complex diseases may be caused by rare, pathogenic variants (“monogenic”) or by the cumulative effect of numerous common variants (“polygenic”). Comprehensive genome interpretation should enable assessment for both monogenic and polygenic components of inherited risk. The traditional approach requires two distinct genetic testing technologies—high coverage sequencing of known genes to detect monogenic variants and a genome-wide genotyping array followed by imputation to calculate genome-wide polygenic scores (GPSs). We assessed the feasibility and accuracy of using low coverage whole genome sequencing (lcWGS) as an alternative to genotyping arrays to calculate GPSs.
Methods
First, we performed downsampling and imputation of WGS data from ten individuals to assess concordance with known genotypes. Second, we assessed the correlation between GPSs for 3 common diseases—coronary artery disease (CAD), breast cancer (BC), and atrial fibrillation (AF)—calculated using lcWGS and genotyping array in 184 samples. Third, we assessed concordance of lcWGS-based genotype calls and GPS calculation in 120 individuals with known genotypes, selected to reflect diverse ancestral backgrounds. Fourth, we assessed the relationship between GPSs calculated using lcWGS and disease phenotypes in a cohort of 11,502 individuals of European ancestry.
Results
We found imputation accuracy r2 values of greater than 0.90 for all ten samples—including those of African and Ashkenazi Jewish ancestry—with lcWGS data at 0.5×. GPSs calculated using lcWGS and genotyping array followed by imputation in 184 individuals were highly correlated for each of the 3 common diseases (r2 = 0.93–0.97) with similar score distributions. Using lcWGS data from 120 individuals of diverse ancestral backgrounds, we found similar results with respect to imputation accuracy and GPS correlations. Finally, we calculated GPSs for CAD, BC, and AF using lcWGS in 11,502 individuals of European ancestry, confirming odds ratios per standard deviation increment ranging 1.28 to 1.59, consistent with previous studies.
Conclusions
lcWGS is an alternative technology to genotyping arrays for common genetic variant assessment and GPS calculation. lcWGS provides comparable imputation accuracy while also overcoming the ascertainment bias inherent to variant selection in genotyping array design.
Background
Cardiovascular disease and cancer are common, complex diseases that remain leading causes of global mortality [1]. Long recognized to be heritable, recent advances in human genetics have led to consideration of DNA-based risk stratification to guide prevention or screening strategies. In some cases, such conditions can be caused by rare, “monogenic” pathogenic variants that lead to a several-fold increased risk—important examples are pathogenic variants in LDLR that cause familial hypercholesterolemia and pathogenic variants in BRCA1 and BRCA2 that underlie hereditary breast and ovarian cancer syndrome. However, the majority of individuals afflicted with these diseases do not harbor any such pathogenic variants. Rather, the inherited susceptibility of many complex traits and diseases is often “polygenic,” driven by the cumulative effect of numerous common variants scattered across the genome [2].
Genome-wide polygenic scores (GPSs) provide a way to integrate information from numerous sites of common variation into a single metric of inherited susceptibility and are now able to identify individuals with a several-fold increased risk of common, complex diseases, including coronary artery disease (CAD), breast cancer (BC), and atrial fibrillation (AF) [3]. For example, for CAD, we previously noted that 8% of the population inherits more than triple the normal risk on the basis of polygenic variation, a prevalence more than 20-fold higher than monogenic familial hypercholesterolemia variants in LDLR that confer similar risk [3].
Comprehensive genome interpretation for common, complex disease therefore could involve both high-fidelity sequencing of important driver genes to identify rare monogenic risk variants and a survey of all common variants across the genome to enable GPS calculation. High coverage whole genome sequencing (hcWGS; for example, 30× coverage) will likely emerge as a single genetic testing strategy, but current prices remain a barrier to large-scale adoption. Instead, the traditional approach has mandated use of two distinct genetic testing technologies—high coverage next generation sequencing (NGS) of important genes to detect pathogenic variants and a genome-wide genotyping array followed by imputation to calculate GPSs.
Low coverage whole genome sequencing (lcWGS; for example, 0.5× coverage) followed by imputation is a potential alternative to genotyping arrays for assessing the common genetic variants needed for GPS calculations. Several recent studies have demonstrated the efficiency and accuracy of lcWGS for other applications of statistical genetics, including local ancestry deconvolution, and complex trait association studies [4,5,6,7].
We developed a pipeline for common genetic variant imputation using lcWGS data on samples from the 1000 Genomes Project (1KGP) [8] and Genome in a Bottle (GIAB) Consortium [9] and herein demonstrate imputation accuracy for lcWGS similar to genotyping arrays. Using three recently published GPSs for CAD [3], BC [10], and AF [3], we show high technical concordance in GPSs calculated from lcWGS and genotyping arrays. Finally, using our pipeline in a large European population seeking genetic testing, we observe similar GPS risk stratification performance as previously published array-based results [3, 10].
Methods
Study design
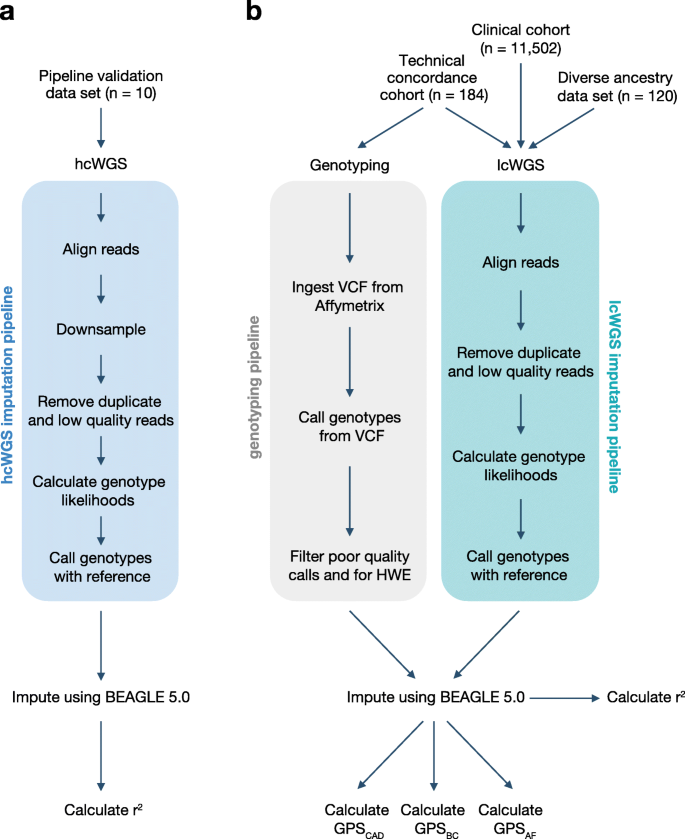
The study design is summarized in Fig. 1 and described in detail below. The pipeline validation data set (n = 10) was used to assess imputation accuracy for common genetic variants (Fig. 1a). The technical concordance cohort (n = 184) was used to assess the correlation between three previously published GPSs for CAD [3], BC [10], and AF [3] from lcWGS and genotyping arrays (Fig. 1b). The diverse ancestry data set (n = 120) was used to assess imputation accuracy for common genetic variants and performance of GPSCAD, GPSBC, and GPSAF (Fig. 1b). The clinical cohort (n = 11,502) was used to assess performance of GPSCAD, GPSBC, and GPSAF in a large European population seeking genetic testing (Fig. 1b).
Study design and imputation pipelines. The study design has four groups: a pipeline validation data set and b technical concordance cohort, diverse ancestry data set, and clinical cohort. The imputation pipeline for each group is depicted. hcWGS, high coverage whole genome sequencing; lcWGS, low coverage whole genome sequencing; HWE, Hardy-Weinberg equilibrium; GPS, genome-wide polygenic score; CAD, coronary artery disease; BC, breast cancer; AF, atrial fibrillation
Data set and cohort selection
The pipeline validation data set included seven globally representative samples from 1KGP populations (HG02155, NA12878, HG00663, HG01485, NA21144, NA20510, and NA19420; Additional file 1: Table S1) [8] and a trio of Ashkenazi samples (NA24385, NA24143, and NA24149) from the GIAB Consortium (Fig. 1a) [9].
The technical concordance cohort included DNA samples from 184 individuals whose healthcare provider had ordered a Color multi-gene panel test (Fig. 1b). All individuals (1) had 85% or greater European genetic ancestry calculated using fastNGSadmix [11] using 1KPG as the reference panel, (2) self-identified as “Caucasian,” and (3) did not have pathogenic or likely pathogenic variants in the multi-gene NGS panel test, as previously described [12] (Additional file 2: Supplementary Methods). Demographics are provided in Additional file 1: Table S2. All phenotypic information was self-reported by the individual through an online, interactive health history tool. Of the 184 individuals, 61 individuals reported having a personal history of CAD (defined here as a myocardial infarction or coronary artery bypass surgery), 62 individuals reported no personal history of CAD, and 61 individuals reported no personal history of CAD but were suspected to have a high GPSCAD based on preliminary analysis. This preliminary analysis included imputation from multi-gene panel and off-target sequencing data, which has been shown to have similar association statistics and effect sizes compared to genotyping arrays [4]. These individuals were included in the technical concordance cohort to artificially create a relatively uniform distribution of GPSCAD in the data set. Correlation coefficients between GPSCAD from lcWGS and genotyping array were calculated after removing the 61 individuals who were suspected to have a high GPSCAD based on multi-gene panel and off-target sequencing data to avoid artificial inflation of the correlation coefficient. Two individuals who reported no personal history of CAD but were suspected to have a high GPSCAD failed genotyping (quality control call rate of < 97%) and lcWGS (overall coverage of < 0.5×), leaving a total of 182 individuals for analyses.
The diverse ancestry data set included a total of 120 samples from the following populations from 1KGP: Han Chinese in Beijing, China (CHB); Yoruba in Ibadan, Nigeria (YRI); Gujarati Indian from Houston, Texas (GIH); Americans of African Ancestry in Southwest USA (ASW); Mexican Ancestry from Los Angeles, USA (MXL); and Puerto Ricans from Puerto Rico (PUR) (Additional file 1: Table S3 and Additional file 3: Figure S1) [8]. Four samples, including NA18917 and NA19147 from the YRI population and NA19729 and NA19785 from the MXL population, were below the target 0.5× coverage and removed from analyses.
The clinical cohort included DNA samples from 11,502 individuals whose healthcare provider had ordered a Color multi-gene panel test (Fig. 1b). All individuals (1) had 90% or greater European genetic ancestry calculated using fastNGSadmix [11] using 1KPG as the reference panel; (2) self-identified as “Caucasian”; (3) provided history of whether they had a clinical diagnosis of CAD, BC, or AF, via an online, interactive health history tool; and (4) did not have pathogenic or likely pathogenic variants detected in the multi-gene sequencing panel test, as previously described [12] (Additional file 2: Supplementary Methods). Demographics are provided in Additional file 1: Table S2.
Whole genome sequencing
DNA was extracted from blood or saliva samples and purified using the Perkin Elmer Chemagic DNA Extraction Kit (Perkin Elmer, Waltham, MA) automated on the Hamilton STAR (Hamilton, Reno, NV) and the Chemagic Liquid Handler (Perkin Elmer, Waltham, MA). The quality and quantity of the extracted DNA were assessed by UV spectroscopy (BioTek, Winooski, VT). High molecular weight genomic DNA was enzymatically fragmented and prepared using the Kapa HyperPlus Library Preparation Kit (Roche Sequencing, Pleasanton, CA) automated on the Hamilton Star liquid handler and uniquely tagged with 10 bp dual-unique barcodes (IDT, Coralville, IA). Libraries were pooled together and loaded onto the NovaSeq 6000 (Illumina, San Diego, CA) for 2 × 150 bp sequencing.
For the pipeline validation data set, all samples underwent WGS with mean coverage of 13.22× (range 7.82× to 17.30×); downsampling was then performed using SAMtools [13] to simulate lcWGS. For the technical concordance cohort, all samples underwent lcWGS with mean coverage of 1.24× (range 0.54× to 1.76×). Imputed genotypes were compared with published, high-confidence known genotypes from 1KGP [8] and the GIAB Consortium [9]. For the diverse ancestry data set, all samples underwent lcWGS with mean coverage of 0.89× (range 0.68× to 1.24×). For the clinical cohort, all samples underwent lcWGS with mean coverage of 0.95× (range 0.51× to 2.57×).
Downsampling
For the pipeline validation data set, aligned reads were downsampled using SAMtools [13] to 2.0×, 1.0×, 0.75×, 0.5×, 0.4×, 0.25×, and 0.1× coverage. For the technical concordance cohort, aligned reads were downsampled to 1.0×, 0.75×, 0.5×, 0.4×, 0.25×, and 0.1× coverage. In a few cases in the technical concordance cohort, the primary samples had fewer reads than the target downsample. In those situations, all of the reads were retained. For example, if the primary sample only had 0.8× coverage, when downsampled to 1.0×, all reads were retained. Downsampling was repeated using two independent seeds in SAMtools. Once the downsampled data was generated, the imputation was repeated to generate imputed genotypes using only the downsampled reads.
Imputation site selection
All data sets and cohorts were imputed to a set of autosomal SNP and insertion-deletion (indel) sites from 1KGP with greater than 1% allele frequency in any of the five 1KGP super populations (African, American, East Asian, European, and South Asian) [8], for a total of 21,770,397 sites. This is hereafter referred to as the “imputation SNP loci.” Multi-allelic SNPs and indels were represented as two biallelic markers for imputation.
Genotype likelihood calculations and imputation
Genotype likelihood calculations and imputation were performed independently for each sample. Sequence reads were aligned with the human genome reference GRCh37.p12 using the Burrows-Wheeler Aligner (BWA) [14], and duplicate and low quality reads were removed. Genotype likelihoods were then calculated at each of the biallelic SNP loci in the imputation SNP loci that were covered by one or more sequencing reads called using the mpileup command implemented in bcftools version 1.8 [15]. Indels or multi-allelic sites were not included in this first genotype likelihood calculation. Reads with a minimum mapping alignment quality of 10 or greater and bases with a minimum base quality of 10 or greater were included. Genotype likelihoods at each observed site were then calculated using the bcftools call command with allele information corresponding to the imputation SNP loci. This procedure discarded calls with indels or calls where the observed base did not match either the reference or the expected alternate allele for the SNP locus.
To convert genotype likelihoods into genotype calls at all imputation SNP loci, two distinct calculations were performed. First, genotypes at imputation SNP loci covered by at least one read were inferred. Genotype calling was performed using the genotype likelihood option implemented in BEAGLE 4.1 [14]. This step is a reference-aware genotype calling step and produces posterior probabilities of genotypes only at sites with at least one read. This algorithm is implemented only in BEAGLE 4.1 [16]. This inference used default BEAGLE 4.1 [16] parameters except with a model scale parameter of 2 and the number of phasing iterations to 0. A custom reference panel was constructed for each sample being imputed by selecting the 250 most similar samples to that sample from 1KGP Phase 3 [8] release using Identity-by-State (IBS) comparison. A reference panel size of 250 was selected to best balance imputation run time and accuracy (Additional file 3: Figure S1). To ensure that IBS values were comparable across samples, a set of regions consistently sequenced at high depth (> 20×) across all samples was utilized. Inclusion of related samples in an imputation reference panel can artificially increase imputation accuracy; therefore, when imputation was performed on samples included in 1KGP Phase 3 release, that sample and any first and second degree related samples (as inferred by the 1KGP data release using genetic data) were excluded from the custom reference panel.
To generate genotypes at all of the remaining untyped sites, a second round of imputation was performed using BEAGLE 5.0 [16]. This imputation used default settings and included the full 1KGP as the imputation reference panel [8]. To note, when performing analysis using 1KGP samples [8], any related individuals were removed. Each sample then had imputed genotype calls at each of the imputation SNP loci. Indels and multi-allelic sites were included in this second genotype likelihood calculation.
Genotyping array
DNA was extracted from blood or saliva samples and purified using the Perkin Elmer Chemagic DNA Extraction Kit (Perkin Elmer, Waltham, MA) automated on the Hamilton STAR (Hamilton, Reno, NV) and the Chemagic Liquid Handler (Perkin Elmer, Waltham, MA). The quality and quantity of the extracted DNA were assessed by UV spectroscopy (BioTek, Winooski, VT).
DNA was genotyped on the Axiom UK Biobank Array by Affymetrix (Santa Clara, CA). Genotypes were filtered according to the manufacturer’s recommendations, removing loci with greater than 5% global missingness and those that significantly deviated from the Hardy-Weinberg equilibrium. In addition, all A/T and G/C SNPs were removed due to potential strand inconsistencies. After applying the above quality filtering and filtering for ambiguous SNP sites, 748,187 SNPs out of an original 830,115 polymorphic sites remained. Each of the remaining SNP orientation was aligned with the hg19 reference sequence to correctly code the reference alleles as allele 1, matching the sequencing data.
To generate genotypes at all of the remaining untyped sites, imputation was performed using BEAGLE 5.0 [16]. This imputation used default settings and included the full 1KGP as the imputation reference panel [8]. To note, when performing analysis using 1KGP samples, any related individuals were removed. Each sample then had imputed genotype calls at each of the imputation SNP loci.
Imputation accuracy and quality assessment
Imputation accuracy for 1KGP and GIAB samples was calculated by comparing imputation results with previously released genotypes, excluding regions marked as low confidence by GIAB.
Imputation accuracy on the genotyped samples was assessed on 470,363 sites that were included in the genotyping array and in the imputation SNP loci at different allele frequency buckets: 257,362 sites with greater than 5% allele frequency, 119,978 sites between 1 and 5% allele frequency, and 93,022 sites with less than 1% allele frequency. Imputation quality was assessed through site-specific dosage r2 comparing with genotype values from the genotyping array.
GPS selection
The GPSs for CAD [3], BC [10], and AF [3] were previously published and selected based on their demonstrated ability to accurately predict and stratify disease risk as well as identify individuals at risk comparable to monogenic disease. GPSCAD contained 6,630,150 polymorphisms, GPSBC contained 3820 polymorphisms, and GPSAF contained 6,730,541 polymorphisms. All loci included in these scores were included in the imputation SNP loci.
GPS normalization
In the clinical cohort, raw GPSs were normalized by taking the standardized residual of the predicted score after correction for the first 10 principal components (PCs) of ancestry [17]. PCs were calculated by projecting lcWGS samples into 10 dimensional PC analyses (PCAs) space using the LASER program [18]. A combination of samples from 1KGP [8] and the Human Origins [19] project were used as a reference for the projection.
Results
Development and validation of imputation pipeline for lcWGS
Previous studies have evaluated the potential use of lcWGS in local ancestry deconvolution, complex trait association studies, and detection of rare genetic variants [4,5,6]. To assess the feasibility and accuracy of this approach for GPSs, we first developed an imputation pipeline that reads raw fastq sequence data and generates a vcf with imputed site information at 21.7 million sites (imputation SNP loci) (Fig. 1a, b). Briefly, reads are aligned to the reference genome and filtered for duplicates and low quality. Using this BAM file, we then calculate genotype likelihoods and impute expected genotypes using 1KGP as the imputation reference panel.
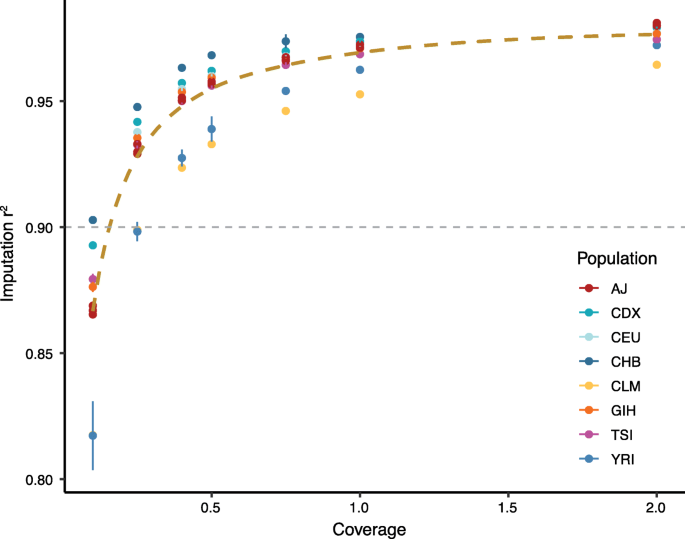
To validate this imputation pipeline, we performed hcWGS and downsampling on seven samples from different 1KGP populations and a trio of Ashkenazi Jewish GIAB samples (pipeline validation data set) to varying depths of coverage from 2.0× to 0.1× (Additional file 1: Table S1) [9]. We used the published genotype calls for each of these samples at all 21 million imputation SNP loci as truth data and found that imputation accuracy was above 0.90 r2 for all samples at 0.5× and higher (Fig. 2). As expected, this was correlated with sequencing depth, with diminishing gains observed at coverages above 1.0×. While imputation accuracy was similar across diverse populations, it was slightly reduced in the Colombian sample (HG01485), likely due to complex local ancestry related to admixture, and in the Yoruban sample (NA19240), likely due to the shorter blocks of linkage disequilibrium and higher genetic diversity in Africa [8]. Taken together, these data suggest that at sequencing depth at or above 0.5×, our pipeline has similar imputation accuracy to genotyping array-based imputation across individuals from multiple populations. As such, we set 0.5× as a quality control for success and removed samples with coverage below this threshold in subsequent analyses.
Assessment of imputation performance in the pipeline validation data set. Downsampling from 30× to 0.1× showed that lcWGS accuracy was above 0.90 r2 for all samples at 0.5× (n = 4 independent random seeds for each sample and coverage value; error bars are 95% confidence intervals). The thick brown dashed line is a smoothed trendline of the average imputation quality while the thin gray dashed line demonstrates previously reported imputation quality from a genotyping array (r2 = 0.90) [4]. AJ, Ashkenazi Jewish; CDX, Chinese Dai in Xishuangbanna, China; CEU, Utah residents with Northern and Western European ancestry; CHB, Han Chinese in Beijing, China; CLM, Colombians from Medellin, Colombia; GIH, Gujarati Indian from Houston, Texas; TSI, Toscani in Italia; YRI, Yoruba in Ibadan, Nigeria
Technical concordance between GPSs calculated from lcWGS and genotyping array
To assess the technical concordance of using lcWGS to calculate GPSs, we performed low coverage sequencing and used genotyping arrays on DNA from 184 individuals (technical concordance cohort) (Fig. 1b). This concordance assessment was restricted to individuals of European ancestry to most closely align with the populations used for GPS training and validation.
We first compared the lcWGS genotype dosages with a subset of variants directly genotyped (n = 470,362) on the genotyping array to assess imputation performance. Assuming the typed loci called on the genotyping array as “true,” we observed an average imputation r2 > 0.90 at 0.5× depth for variants with global minor allele frequency (MAF) greater than 5% and for variants with European MAF greater than 5% (Additional file 3: Figure S2). As expected, imputation accuracy was highest for variants with higher MAF. For lower frequency variants, we saw a reduction in imputation accuracy, as expected, with r2 > 0.85 for variants at 1 to 5% MAF and r2 > 0.80 for variants less than 1% global MAF. Taken together, this demonstrates that lcWGS has high accuracy in this test setting.
We then calculated previously published GPSs for CAD [3], BC [10], and AF [3] on each sample using genotyping array data or lcWGS data. We found that GPSCAD, GPSBC, and GPSAF were highly correlated (Fig. 3a–c), with the score mean (Student’s t test p = 0.17) and variance (F test p = 0.91) equivalent between lcWGS and the genotyping array. The correlations of GPSCAD and GPSAF (r2 = 0.98 and r2 = 0.97, respectively) were slightly higher than that of GPSBC (r2 = 0.93). There are a few key distinctions between the GPSBC and the GPSAF and GPSCAD, which could be responsible for these differences. These include (1) the smaller number of variants in GPSBC (3820 versus 6.6 million), (2) differences in allele frequencies between SNPs with high weights, and (3) GPSBC which was trained and validated on a different genotyping array, the OncoArray, than the Axiom UK Biobank Array used in this study [10]. To match published scores, GPS calculation included all variants regardless of imputation quality. We observed no strong differences in the distribution of observed dosages at GPS loci between lcWGS and genotyping array (Additional file 3: Figure S3). In addition, we found no difference in average difference rates for all three GPSs between blood- and saliva-derived DNA samples using lcWGS (p = 0.53 for CAD, p = 0.21 for BC, p = 0.70 for AF, Additional file 3: Figure S4). We also found no differences in imputation accuracy at variants with MAF > 5% (p = 0.23), variants with MAF between 1 and 5% (p = 0.13), and variants with MAF < 1% (p = 0.07). This is similar to previous results that have demonstrated no differences in error profiles once coverage variability is accounted for [20].
Correlation of GPSs between genotyping array and lcWGS in the technical concordance cohort. a GPSCAD calculated using lcWGS was highly correlated (r2 = 0.98) with those calculated using genotyping array (n = 182). b GPSBC calculated using lcWGS was highly correlated (r2 = 0.93) with those calculated using genotyping array (n = 182). c GPSAF was highly correlated (r2 = 0.97) with those calculated using genotyping arrays (n = 182). x-axis is the raw GPS calculated from the genotyping array, and y-axis is the raw GPS calculated from the lcWGS data; raw GPS values are unitless. lcWGS low coverage whole genome sequencing; GPS, genome-wide polygenic score; CAD, coronary artery disease; BC, breast cancer; AF, atrial fibrillation
The technical concordance cohort ranged in coverage from 0.54× to 1.76× with a mean coverage of 1.24×, and we have shown that depth can impact imputation performance—depth increases above 0.5× have a smaller but measurable effect on imputation performance (Fig. 2; Additional file 3: Figure S2). To determine the low coverage sequencing depth required for GPS accuracy, we used SAMtools [13] to downsample the lcWGS data in this cohort to 1.0×, 0.75×, 0.5×, 0.4×, 0.25×, and 0.1×. We found that GPSCAD, GPSBC, and GPSAF are robust to lcWGS sequencing depth 0.5× and that coverages do not systematically bias GPS calculations in a specific direction (Additional file 3: Figures S5, S6, and S7). Interestingly, correlation at 0.1× was still high enough that GPSs at this coverage may have research utility, suggesting that significant amounts of data regarding common genetic variation could be recovered from off-target reads in exome and multi-gene panel sequencing studies to allow for GPS calculation. Taken together, these data demonstrate that lcWGS provides equivalent accuracy for calculation of GPSs, with sequencing coverage as low as 0.5×.
Assessment of imputation performance and technical concordance across diverse populations
To further assess the performance of our imputation pipeline across diverse populations, we performed lcWGS on 120 additional samples from 6 1KGP populations (CHB, GIH, YRI, ASW, MXL, and PUR; Additional file 1: Table S3) that represent the range of ancestry observed in admixed populations (diverse ancestry data set) [8]. We compared genotypes imputed using our lcWGS pipeline to known 1KGP WGS data at all 21 million imputation SNP loci and found that imputation accuracy was above 0.90 r2 for all samples (range 0.94–0.97) (Fig. 4a). In addition, we found that GPS calculated from lcWGS data and GPS calculated from the Phase 3 1KGP WGS data release have a high correlation, with an r2 value of 0.98, 0.91, and 0.98 for CAD, BC, and AF, respectively (Fig. 4b–d). These results suggest that lcWGS can enable accurate imputation and calculation of GPSs in diverse populations.
Assessment of imputation performance and technical concordance across diverse populations. a Imputed genotypes calculated using lcWGS data were highly correlated with genotypes from known 1KGP data (n = 116), with all samples having an imputation quality above 0.90 r2. The thin gray dashed line demonstrates previously reported imputation quality from a genotyping array (r2 = 0.90) [4]. b GPSCAD calculated using lcWGS data was highly correlated (r2 = 0.98) with those calculated using known 1KGP data (n = 116). c GPSBC calculated using lcWGS data was highly correlated (r2 = 0.91) with those calculated using known 1KGP data (n = 116). d GPSAF was highly correlated (r2 = 0.98) with those calculated using known 1KGP data (n = 116). 1KGP, 1000 Genomes Project; lcWGS, low coverage whole genome sequencing; GPS, genome-wide polygenic score; CAD, coronary artery disease; BC, breast cancer; AF, atrial fibrillation
Association of lcWGS-calculated GPSs with disease phenotypes in a clinical cohort
Previous studies have demonstrated the association of GPSs with prevalent disease using genotyping arrays [3, 10, 21,22,23] and hcWGS [17]. To observe the performance of lcWGS-calculated GPSs in a large population, we performed low coverage sequencing on 11,502 European individuals (clinical cohort) (Additional file 1: Table S2) and calculated GPSCAD, GPSBC, and GPSAF for each individual. Raw GPSs were normalized by taking the standardized residual of the predicted score after correction for the first 10 PCAs (Additional file 3: Figure S8) [17, 24]. First, we note that there are no major outliers (defined as a z-score greater than 5) in GPSCAD, GPSBC, and GPSAF and that the normalized scores formed an approximately normal distribution for each (Additional file 3: Figure S9). Each of the GPSs was strongly associated with self-reported history of disease, with effect estimates comparable to prior reports using genotyping arrays to calculate GPS—GPSCAD (OR per standard deviation = 1.59 (1.32–1.92); n = 11,010), GPSBC (OR per standard deviation = 1.56 (1.45–1.68); n = 8722), and GPSAF (OR per standard deviation = 1.28 (1.12–1.46); n = 10,303) (Fig. 5).
Association of lcWGS-calculated GPSs with disease phenotypes in the clinical cohort. lcWGS-calculated GPSCAD was associated with personal history of CAD (OR = 1.589 (1.32–1.92), n = 11,010, p = 1.32 × 10−6). GPSCAD was adjusted for age and sex. lcWGS-calculated GPSBC was associated with personal history of BC (OR = 1.56 (1.45–1.68); n = 8722, p = 1.0 × 10−16). GPSBC was calculated only for females and adjusted for age at menarche. lcWGS-calculated GPSAF was associated with personal history of AF (OR = 1.277 (1.12–1.46); n = 10,303, p = 0.000292). GPSAF was adjusted for age and sex. lcWGS, low coverage whole genome sequencing; GPS, genome-wide polygenic score; CAD, coronary artery disease; BC, breast cancer; AF, atrial fibrillation
Area under the curve (AUC) is an additional metric used to assess the ability of a given risk factor to discriminate between affected cases and disease-free controls. When only the GPS was included in the prediction model, GPSCAD had an AUC of 0.60, GPSBC had an AUC of 0.63, and GPSAF had an AUC of 0.57. The additional inclusion of age and sex increased the AUCs to 0.86 for GPSCAD, 0.78 for GPSBC, and 0.78 for GPSAF. For each of these three diseases, the magnitude of associations with clinical disease and AUC metrics was consistent with previous publications [3, 10]. Taken together, these results suggest that lcWGS-calculated GPSs can accurately stratify risk with comparable accuracy to previously published GPS-disease associations calculated on the basis of genotyping array data.
Discussion
For the past two decades, genotyping array-based GWAS and imputation have been the driving force in our discovery of genetic loci predictive of disease and derivation and calculation of GPSs. In this study, we developed and validated an imputation pipeline to calculate GPSs from variably downsampled hcWGS and lcWGS data sets. While the efficiency of lcWGS has been reported for other applications of statistical genetics [4,5,6], we demonstrate that lcWGS achieves similar technical concordance as the Axiom UK Biobank Array by Affymetrix for determining GPSs. Furthermore, the imputation r2 from lcWGS was greater than 90%, which is similar to the imputation accuracy reported from other commercially available genotyping arrays [25]. Taken together, these data suggest that lcWGS has comparable accuracy to genotyping arrays for assessment of common variants and subsequent calculation of GPSs.
Our finding that lcWGS can be used for accurate genotyping and imputation of common genetic variants has implications for the future of genomic research and medicine. Currently, disease GWAS are performed using a variety of genotyping arrays designed to target specific sets of genes or features, reducing imputation quality in regions that are not targeted [26]. lcWGS enables less biased imputation than genotyping arrays by not pre-specifying the genetic content that is included for assessment, as is necessary for genotyping arrays. Because initial GWAS focused on populations with high homogeneity to reduce noise and increase fit of risk stratification, many genotyping arrays were designed to capture common genetic variants based on the linkage disequilibrium structure in European populations [27]. However, this ascertainment bias reduces the imputation performance from genotyping array data in diverse populations [28,29,30]. Imputation from lcWGS data reduces this bias by including all SNPs observed in 1KGP populations as potential predictors. The effects of SNP selection bias are also not equivalent across genotyping arrays, and therefore, variants included in a GPS trained and validated on one genotyping array may not be as predictive on another genotyping array [31]. lcWGS systematically surveys variants independent of SNP selection bias and thus provides one approach to overcome this issue. Our findings here demonstrate that GPSs trained and validated on different genotyping arrays are transferable to lcWGS-calculated GPS. Furthermore, as new genetic associations are discovered, lcWGS can be re-analyzed with ever more inclusive sets of identified. By contrast, genotyping arrays are static and cannot be easily updated or changed without designing a de novo platform.
lcWGS also has the potential to easily integrate into current clinical sequencing pipelines. In contrast to genotyping arrays, which require investment in separate laboratory technology, lcWGS can be performed on the same platform as current hcWGS or targeted multi-gene panel clinical testing. The ease of combining these two pathways could help to drive GPS adoption into clinical practice and can likely be achieved at a cost comparable to genotyping arrays [4]. In addition, lcWGS could be used to detect large insertions and deletions.
This study should be interpreted in the context of potential limitations. First, the imputation accuracy observed in our analysis may have been limited by the reference panel size. Future efforts using an even larger reference or more diverse panel may lead to further improved imputation accuracy, particularly for variants with allele frequency less than 1% [25, 32]. Second, while lcWGS may ultimately enable derivation of GPSs with improved predictive accuracy or ethnic transferability, this was not explicitly explored here. Rather, we demonstrate the feasibility and accuracy of using lcWGS of calculating GPSs published in previous studies. Third, disease phenotypes in our clinical cohort were based on individual self-report rather than review of health records. However, several studies have shown that self-reported personal history data have high concordance with data reported by a healthcare provider or electronic health records [33,34,35,36], and any inaccuracies would be expected to bias GPS-disease associations to the null. Fourth, while lcWGS data provides accurate inference of common variants, imputation is less accurate for rare variants. High coverage clinical sequencing of genes, such as those in the American College of Medical Genetics and Genomics (ACMG) list of genes in which pathogenic variants are deemed important and actionable [33], is essential for accurate detection of rare pathogenic variants.
Conclusions
In conclusion, this work establishes lcWGS as an alternative approach to genotyping arrays for common genetic variant assessment and GPS calculation—providing comparable accuracy at similar cost while also overcoming the ascertainment bias inherent to variant selection in genotyping array design.
Availability of data and materials
The technical concordance and clinical cohort data are not publicly available because the research participant consent, privacy policy, and terms of service of the commercial laboratory do not include authorization to share data.
1KGP [8], http://www.internationalgenome.org/
GIAB [9], ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/
Samtools/Bcftools [13], http://www.htslib.org/
BEAGLE [16], https://faculty.washington.edu/browning/beagle/beagle.html
FastNGSAdmix [11], http://www.popgen.dk/software/index.php/FastNGSadmix
Abbreviations
- GPS:
-
Genome-wide polygenic score
- lcWGS:
-
Low coverage whole genome sequencing
- CAD:
-
Coronary artery disease
- BC:
-
Breast cancer
- AF:
-
Atrial fibrillation
- 1KGP:
-
1000 Genomes Project
- GIAB:
-
Genome in a Bottle
- Indel:
-
Insertion-deletion
- BWA:
-
Burrows-Wheeler Aligner
- IBS:
-
Identity-by-State
- PCs:
-
Principal components
- PCA:
-
PC analysis
- MAF:
-
Minor allele frequency
- AUC:
-
Area under the curve
References
Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128.
Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86.
Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018; Available from: https://doi.org/10.1038/s41588-018-0183-z.
Pasaniuc B, Rohland N, McLaren PJ, Garimella K, Zaitlen N, Li H, et al. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat Genet. 2012;44:631–5.
Gilly A, Southam L, Suveges D, Kuchenbaecker K, Moore R, Melloni GEM, et al. Very low depth whole genome sequencing in complex trait association studies. Bioinformatics. 2018; Available from: https://doi.org/10.1093/bioinformatics/bty1032.
Liu S, Huang S, Chen F, Zhao L, Yuan Y, Francis SS, et al. Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and Chinese population history. Cell. 2018;175:347–59 e14.
Navon O, Sul JH, Han B, Conde L, Bracci PM, Riby J, et al. Rare variant association testing under low-coverage sequencing. Genetics. 2013;194:769–79.
1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Zook JM, Catoe D, McDaniel J, Vang L, Spies N, Sidow A, et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025.
Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104:21–34.
Jørsboe E, Hanghøj K, Albrechtsen A. fastNGSadmix: admixture proportions and principal component analysis of a single NGS sample. Bioinformatics. 2017;33:3148–50.
Neben CL, Zimmer AD, Stedden W, van den Akker J, O’Connor R, Chan RC, et al. Multi-gene panel testing of 23,179 individuals for hereditary cancer risk identifies pathogenic variant carriers missed by current genetic testing guidelines. J Mol Diagn. 2019. Elsevier; [cited 2019 Jun 11];0. Available from: https://jmd.amjpathol.org/article/S1525-1578(18)30334-9/fulltext.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN]. 2013; Available from: http://arxiv.org/abs/1303.3997.
Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93.
Browning BL, Zhou Y, Browning SR. A one-penny imputed genome from next-generation reference panels. Am J Hum Genet. 2018;103:338–48.
Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, et al. Whole genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation. 2018; American Heart Association Bethesda, MD; [cited 2018 Nov 27]; Available from: https://www.ahajournals.org/doi/abs/10.1161/CIRCULATIONAHA.118.035658.
Wang C, Zhan X, Liang L, Abecasis GR, Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet. 2015;96:926–37.
Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–24.
Wall JD, Tang LF, Zerbe B, Kvale MN, Kwok P-Y, Schaefer C, et al. Estimating genotype error rates from high-coverage next-generation sequence data. Genome Res. 2014;24:1734–9.
Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol. 2018;72:1883–93.
Richardson TG, Harrison S, Hemani G, Smith GD. An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. bioRxiv. 2018:467910 [cited 2018 Nov 27]. Available from: https://www.biorxiv.org/content/early/2018/11/11/467910.
Mavaddat N, Pharoah PDP, Michailidou K, Tyrer J, Brook MN, Bolla MK, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107 Available from: https://doi.org/10.1093/jnci/djv036.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83.
Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793.
Lachance J, Tishkoff SA. SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it. Bioessays. 2013;35:780–6.
Wojcik GL, Fuchsberger C, Taliun D, Welch R, Martin AR, Shringarpure S, et al. Imputation-aware tag SNP selection to improve power for large-scale, multi-ethnic association studies. G3. 2018;8:3255–67.
Nelson SC, Doheny KF, Pugh EW, Romm JM, Ling H, Laurie CA, et al. Imputation-based genomic coverage assessments of current human genotyping arrays. G3. 2013:1795–807 Available from: https://doi.org/10.1534/g3.113.007161.
Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S, et al. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol. 2013;11:e1001661.
Johnson EO, Hancock DB, Levy JL, Gaddis NC, Saccone NL, Bierut LJ, et al. Imputation across genotyping arrays for genome-wide association studies: assessment of bias and a correction strategy. Hum Genet. 2013;132:509–22.
Kowalski MH, Qian H, Hou Z, Rosen JD, Tapia AL, Shan Y, et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. bioRxiv. 2019:683201 [cited 2019 Oct 1]. Available from: https://www.biorxiv.org/content/10.1101/683201v1.
Gentry-Maharaj A, Fourkala E-O, Burnell M, Ryan A, Apostolidou S, Habib M, et al. Concordance of National Cancer Registration with self-reported breast, bowel and lung cancer in England and Wales: a prospective cohort study within the UK Collaborative Trial of Ovarian Cancer Screening. Br J Cancer. 2013;109:2875–9.
D’Aloisio AA, Nichols HB, Hodgson ME, Deming-Halverson SL, Sandler DP. Validity of self-reported breast cancer characteristics in a nationwide cohort of women with a family history of breast cancer. BMC Cancer. 2017;17:692.
Kehoe R, Wu SY, Leske MC, Chylack LT Jr. Comparing self-reported and physician-reported medical history. Am J Epidemiol. 1994;139:813–8.
Malmo V, Langhammer A, Bønaa KH, Loennechen JP, Ellekjaer H. Validation of self-reported and hospital-diagnosed atrial fibrillation: the HUNT study. Clin Epidemiol. 2016;8:185–93.
Acknowledgements
We thank Will Stedden, Carmen Lai, and Anjali D. Zimmer for helpful discussions and Justin Lock, Alok Sabnis, and Valerie Ngo for laboratory support and sample processing.
Funding
This work was supported by Color Genomics.
Author information
Authors and Affiliations
Contributions
JRH, GM, AYZ, and AVK designed the overall study. JRH, CLN, GM, AYZ, SK, and AVK contributed to the data acquisition and analysis. JRH, CLN, GM, AYZ, SK, and AVK drafted or critically revised the manuscript for important intellectual content. AYZ and AVK are the guarantors of this work and, as such, have full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All individuals in the technical concordance cohort and clinical cohort gave electronic informed consent to have their de-identified information and sample used in anonymized studies (Western Institutional Review Board, #20150716). This research study conformed to the principles of the Helsinki Declaration.
Consent for publication
All individuals in the technical concordance cohort and clinical cohort gave electronic informed consent that Color may author publications using non-aggregated, de-identified information, either on its own or in collaboration with academic or commercial third parties.
Competing interests
JRH, CLN, and AYZ are currently employed by and have equity interest in Color Genomics. JRH has previously consulted for Twist Bioscience and Etalon Diagnostics. GM was previously employed at Color Genomics and Operator. JRH and GM report a patent application related to low coverage whole genome sequencing. SK is an employee of Verve Therapeutics and holds equity in Verve Therapeutics, Maze Therapeutics, Catabasis, and San Therapeutics. He is a member of the scientific advisory boards for Regeneron Genetics Center and Corvidia Therapeutics; he has served as a consultant for Acceleron, Eli Lilly, Novartis, Merck, Novo Nordisk, Novo Ventures, Ionis, Alnylam, Aegerion, Haug Partners, Noble Insights, Leerink Partners, Bayer Healthcare, Illumina, Color Genomics, MedGenome, Quest, and Medscape; he reports patents related to a method of identifying and treating a person having a predisposition to or afflicted with cardiometabolic disease (20180010185) and a genetic risk predictor (20190017119). AVK has served as a consultant for Color Genomics and reports a patent related to a genetic risk predictor (20190017119).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Samples in the pipeline validation data set. Table S2. Demographics of technical concordance cohort and clinical cohort. Table S3. Samples in the diverse ancestry data set.
Additional file 2.
Supplementary Methods.
Additional file 3: Figure S1.
Association of lcWGS time and accuracy for samples in the pipeline validation data set at 1.0X coverage. Figure S2. Imputation performance of the pipeline compared to genotyping array for different allele frequencies. Figure S3. Observed genotype dosages at GPS loci between lcWGS data and genotyping array. Figure S4. Comparison of blood-derived and saliva-derived samples. Figure S5. Correlation of GPSs between genotyping array and lcWGS at different coverage depths in the technical concordance cohort., Figure S6. Correlation of GPSCAD between genotyping array and lcWGS at different coverage depths in the technical concordance cohort when removing individuals who were suspected to have a high GPSCAD. Figure S7. Concordance of GPS calculated at different coverages using different sampling seeds in the technical concordance cohort. Figure S8. First two principal components of ancestry. Figure S9. Distribution of GPSs in the clinical cohort.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Homburger, J.R., Neben, C.L., Mishne, G. et al. Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores. Genome Med 11, 74 (2019). https://doi.org/10.1186/s13073-019-0682-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13073-019-0682-2