- Review
- Open access
- Published:
Best practices for variant calling in clinical sequencing
Genome Medicine volume 12, Article number: 91 (2020)
Abstract
Next-generation sequencing technologies have enabled a dramatic expansion of clinical genetic testing both for inherited conditions and diseases such as cancer. Accurate variant calling in NGS data is a critical step upon which virtually all downstream analysis and interpretation processes rely. Just as NGS technologies have evolved considerably over the past 10 years, so too have the software tools and approaches for detecting sequence variants in clinical samples. In this review, I discuss the current best practices for variant calling in clinical sequencing studies, with a particular emphasis on trio sequencing for inherited disorders and somatic mutation detection in cancer patients. I describe the relative strengths and weaknesses of panel, exome, and whole-genome sequencing for variant detection. Recommended tools and strategies for calling variants of different classes are also provided, along with guidance on variant review, validation, and benchmarking to ensure optimal performance. Although NGS technologies are continually evolving, and new capabilities (such as long-read single-molecule sequencing) are emerging, the “best practice” principles in this review should be relevant to clinical variant calling in the long term.
Background
The emergence of next-generation sequencing more than a decade ago represented a major technological advance over traditional sequencing methods. NGS technologies enabled ambitious large-scale genomic sequencing efforts that have transformed our understanding of human health and disease, such The Cancer Genome Atlas [1,2,3,4,5,6,7,8], the Centers for Mendelian Genomics [9], and the UK10K Project [10]. They have also been widely adopted for clinical genetic testing. Whole-exome sequencing, which selectively targets the protein-coding regions of known genes, has become a frontline diagnostic tool for inherited disorders [11,12,13,14]. Targeted panels which leverage this approach to interrogate medically relevant subsets of genes have become core components of precision oncology [15,16,17].
The characteristics and sheer volume of NGS reads necessitated the development of a new generation of computational algorithms and analysis pipelines equipped to handle such data. As NGS technologies have matured, so too have the software tools for key analytical tasks, such as variant calling. Ten years and thousands of samples later, we now have a much deeper understanding of the capabilities and limitations of NGS for detecting and characterizing sequence variation. In this review, I discuss the current “best practices” for variant calling in clinical sequencing for both germline analysis in family trios and somatic analysis of tumor-normal pairs. This includes recommendations for the choice of sequencing strategy, NGS read alignment/preprocessing, combination of multiple variant calling tools, and rigorous filtering to remove false positives. I also include guidance on benchmarking NGS analysis pipeline performance using “gold standard” reference datasets to achieve the optimum balance of sensitivity and specificity.
Sequencing strategies and implications
The choice of sequencing strategy for a clinical sample has important ramifications for variant calling (Table 1). Single- or multi-gene panels are increasingly cost-effective means of testing for subsets of genes associated with specific clinical phenotypes. For example, the OtoSCOPE hearing loss panel [18] targets 89 genes and microRNAs associated with hearing loss (1574 total exons); across a cohort of 711 sequenced patients, the average sequence depth achieved was 716× per patient. Numerous gene panels are commercially available, ranging in size from a single gene to hundreds of genes. Exome sequencing, which targets virtually all ~ 20,000 protein-coding genes, typically achieves > 100× average depth across the target regions. Whole-genome sequencing offers the most comprehensive approach and typically yields ~ 30–60× average sequence depth across the entire genome. Other considerations, such as cost and turnaround time, also influence the choice of sequencing strategy but are beyond the scope of this review.
These differences in depth and breadth of sequencing coverage have implications on variant calling. All three strategies generally offer excellent sensitivity for detecting SNVs/indels using tools such as GATK HaplotypeCaller [19] and Platypus [20]. Copy number variants (CNVs) spanning multiple exons can be called with reasonable sensitivity using panel and exome data [21]. Whole-genome sequencing remains the superior strategy for the comprehensive detection of all types of sequence variants. However, it should be noted that the higher sequence depth achieved in panel and exome sequencing may enable more sensitive detection of variants at low allele frequencies, e.g., subclonal somatic mutations in cancer and mosaic germline variants [22,23,24].
Alignment and pre-processing
The primary analysis of sequencing data, including its alignment to a reference sequence, is a critical phase of NGS analysis. A selection of recommended tools can be found in the top of Table 2.
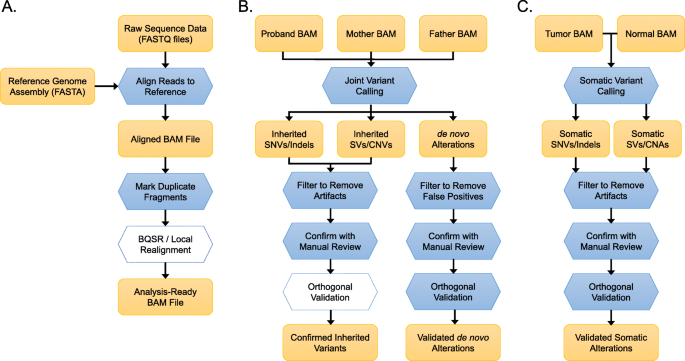
In a typical pipeline (Fig. 1a), raw sequence data in FASTQ format are aligned to the reference sequence using an aligner such as BWA-Mem [25], with the resulting alignments typically stored in binary alignment/map (BAM) file format [31]. Because of their compressed file size, indexed-access capabilities, and standardized data formats, BAM files have become the standard format for storing and sharing NGS data. The Samtools package [31] provides most of the BAM file manipulation tools required for clinical sequencing.
Standard pipelines for NGS analysis. a Alignment and pre-processing of NGS data for an individual sample. Raw sequence data in FASTQ format are aligned to the reference sequence, with the resulting alignments typically stored in binary alignment/map (BAM) file format. Marking of duplicates in the BAM file is a critical step to account for duplicate reads of the same fragment. Base quality score recalibration (BQSR) and local realignment around indels are a computationally expensive step that may marginally improve variant calls. At the conclusion of this step, the file is ready for variant analysis. b Variant calling in NGS trio sequencing. In this common study design, variants are called jointly (simultaneously) in a proband and both parents, which enables the phasing of variants by parent of origin. The initial variant calls are typically filtered to remove a number of recurrent artifacts associated with short-read alignment and maybe visually confirmed by manual review of the sequence alignments. Orthogonal validation may be performed to confirm the variant and its segregation within the family. De novo alterations should be aggressively filtered to remove both artefactual calls in the proband (false positives) and inherited variants that were under-called in a parent (false negatives). In addition to manual inspection of alignments, most de novo mutations are independently verified by orthogonal validation techniques, such as Sanger sequencing. c Somatic variant calling in matched tumor-normal pairs. Identification of somatic alterations in tumors requires specialized variant callers which consider aligned data from the tumor and normal simultaneously. Candidate somatic variants are filtered and visually reviewed to remove common alignment artifacts as well as germline variants under-called in the normal sample. The resulting variants are typically validated by orthogonal approaches, which may require specialized approaches for low-frequency variants
Once NGS data are aligned to the reference sequence, it is possible to identify redundant reads that originated from the same DNA sequence molecule. These “PCR duplicates” represent 5–15% of sequencing reads in a typical exome [64] and can be identified on the basis of the alignment position and read pairing information. Tools such as Picard [28] and Sambamba [29] identify and mark duplicate reads in a BAM file to exclude them from downstream analysis.
The GATK Best Practices workflow [65] recommends two additional steps for pre-processing BAM files prior to variant calling. The first is base quality score recalibration (BQSR), which adjusts the base quality scores of sequencing reads using an empirical error model. The second is local realignment around indels, which aims to reduce false-positive variant calls caused by alignment artifacts (discussed below). Evaluations of variant calling accuracy before and after BQSR/realignment suggest that the improvements are marginal [66]; because of this and the high computational cost, this may be viewed as an optional step for pre-processing.
Routine quality control (QC) of analysis-ready BAMs should be performed prior to variant calling to evaluate key sequencing metrics [28], to verify that sufficient sequencing coverage was achieved [32], and to check samples for evidence of contamination [35]. In the case of family studies and paired samples (e.g., tumor-normal), expected sample relationships should be confirmed with tools for relationship inference such as the KING algorithm [34].
Benchmarking resources for variant calling
Evaluating the accuracy of variant calls requires access to benchmark datasets in which the true variants are already known. Several such benchmarking resources have been made publicly available in recent years. The most widely used ones include the Genome in a Bottle (GIAB) [67] and the Platinum Genome [68] datasets for NA12878, a human sample of European ancestry that has been sequenced with various technologies at laboratories around the world. Each benchmarking dataset includes a set of “ground truth” small variant calls (SNVs and indels) based on the consensus of several variant calling tools, as well as defining the “high-confidence” regions of the human genomes in which variant calls can be benchmarked against a variety of public resources. The GIAB dataset has been continually improved with the addition of data from multiple short-read and linked-read sequencing datasets and the expansion of the reference from one sample to seven [69]. The Global Alliance for Genomics and Health has also established a best practice framework to guide evaluations of variant calling accuracy using these resources [70]. As discussed in this paper, sophisticated comparison tools which account for subtle differences in variant representation are recommended when comparing a set of variant calls against a benchmark resource.
One drawback of the aforementioned benchmarking resources is that many of the same sequencing technologies and variant calling algorithms evaluated against them were also used to construct the reference datasets in the first place. Synthetically created datasets in which the positions of all sequence variants are known a priori have been published to address this issue. For example, the synthetic diploid (Syndip) dataset is derived from de novo long-read assemblies of two homozygous human cell lines and aims to provide a less biased view of variant calling accuracy genome-wide [71]. Syndip is uniquely advantaged to provide benchmarking data for more challenging regions of the genome, such as duplicated sequences. Although the cell lines themselves are not in a public repository, sequencing datasets for both are widely available. More guidance on using benchmarking datasets to optimize variant calling performance is offered in the relevant sections below.
Best practices for germline variant calling
Dozens of variant calling tools for NGS data have been published in the past 10 years, and countless more have been developed by researchers for internal use. A selection of exemplar tools grouped by purpose can be found in the middle of Table 2. Because SNV/indel detection tools such as GATK HaplotypeCaller have demonstrated high accuracy (F-scores > 0.99) in numerous benchmark datasets, choosing a single variant caller that meets the needs of the laboratory (in terms of pipeline compatibility and ease of implementation) is usually sufficient. However, combining the results of two orthogonal SNV/indel callers, such as HaplotypeCaller and Platypus, may offer a slight sensitivity advantage. Software packages such as BCFtools make it possible to merge and reconcile multiple variant callsets (in VCF format) into one, though care should be taken to properly handle complex variants and/or differences in variant representation [70].
To discuss the recommended best practices for germline variant calling, we will consider trio sequencing for inherited disorders, which is a common scenario for clinical genetic testing. A trio analysis pipeline typically begins with the analysis-ready BAM files for the proband and both parents (Fig. 1b). For optimal results, all three samples should be sequenced under identical protocols (capture kit, instrument, and reagent kit) and processed with identical alignment and pre-processing steps. This is particularly important for copy number variant calling and SV calling, which rely on uniform sequencing depth and library insert size, respectively.
Individual versus joint variant calling
Virtually, all variant calling tools can be applied to individual samples after alignment and pre-processing are complete. It may be preferable, therefore, to perform variant calling on every sample as it comes through the pipeline. Doing so can facilitate automation of NGS analysis, which may be desirable for laboratories processing large numbers of samples. Individual VCF files can be merged later using BCFtools or similar packages; however, it should be noted that VCF files typically only contain entries for positions that are variant in a particular sample. In other words, when a variant is only detected in some samples but not others, it is not clear whether the other samples are wild type for that position or simply did not achieve sufficient coverage for the variant caller to make a call.
Joint variant calling—which considers all samples simultaneously—offers several key advantages. First, it produces called genotypes for every sample at all variant positions, not just the ones that were detected in a given individual. This makes it possible to differentiate between a position that matches the reference sequence with high probability and a position in which the sample did not achieve sufficient coverage. Second, in the case of trio sequencing, joint calling enables direct inference of phase information to establish, for example, whether two heterozygous variants in a proband are in cis or in trans. Third, it mitigates the issue of variant representation differences which might otherwise be problematic, particularly for complex variants [72]. Finally, joint analysis allows a variant caller to use information from one sample to infer the most likely genotype in another, which has been shown to increase the sensitivity of variant calling in low-coverage regions [19].
SNV/indel calling
Numerous tools have been developed to identify single nucleotide variants (SNVs) and short insertions/deletions (indels) from aligned NGS data. Most tools for this purpose, such as Samtools/BCFtools [37] and FreeBayes [36], employ Bayesian statistics to infer the most likely genotype. GATK HaplotypeCaller [19] and Platypus [20] also employ local realignment or assembly of sequencing reads to improve the accuracy of variant calls. Numerous studies have compared the relative performance of these tools on various datasets and have found, generally, that they produce similar results: variant concordance is typically 80–90% concordance or higher, with most differences are attributed to variants at low-coverage or low-confidence positions [73,74,75,76]. Even so, such differences could amount to thousands of variant calls genome-wide. Thus, it is important not only to choose a robust variant caller for SNVs/indels, but also to benchmark and fine-tune it to achieve optimal performance on the data to be analyzed.
Filtering to remove artifacts
The accuracy of NGS variant calls relative to the previous “gold standard” of Sanger sequencing has been well documented at > 99% [77,78,79]. However, it should be noted that NGS data are prone to certain types of artifactual variant calls, many of which are related to errors in short-read alignment [37, 66]. Numerous groups including ours have investigated the source of artifacts and demonstrated that they can be systematically filtered without significantly compromising sensitivity [41, 44]. Even so, visual review of the alignments for clinically relevant variants, using a tool like the Integrative Genomics Viewer [63], is recommended to identify false-positive variant calls that slip past automated filters.
Figure 2 depicts several frequently occurring artifacts that can be identified by manual review: low-quality base calls (Fig. 2a), read-end artifacts (Fig. 2b) due to local misalignment near indels (Fig. 2c), strand bias artifacts (Fig. 2d), erroneous alignments in low-complexity regions (Fig. 2e), and paralogous alignments of reads not well represented in the reference (Fig. 2f).
Common artifacts in NGS alignments that gave rise to a false-positive de novo mutation call in a family trio. Each pane is an IGV screenshot of WGS alignments for the proband (top track), mother, (middle track), and father (bottom track). Each sample’s track comprises two parts: a histogram of the read depth and the reads as aligned to the reference sequence. Reads are colored according to the aligned strand (red = forward strand; blue = reverse strand). a False positive associated with low base quality. Most reads supporting the variant have low base quality indicated by lightly shaded non-reference bases. Four reads in the proband showed the alternate allele with good quality, triggering the variant call. b False positive due to misalignments near the start or end of reads. Notice that the alternate allele is only observed at the start/end of reads in the proband. In this case, the read depth histogram provides a clue as to the cause of the misalignment. As shown in the next panel, this occurs at the breakpoint of a large paternally inherited deletion. c The same position as in b, but with soft-clipped bases shown in color. BLAT alignment of such reads reveals that the soft-clipped portion matches the other side of the deletion segment some 5.2 kb downstream. d False positive associated with strand bias. All but one variant-supporting reads in the proband are on the reverse strand, whereas reference-supporting reads are equally represented on both strands. e False positives associated with low-complexity sequences. In this case, reads erroneously showing a single-base deletion (horizontal black line) at a T-homopolymer are enriched in the proband. R supporting insertions (purple) are also seen. Note that this position is zoomed out compared to the other panels, a recommended practice to visualize the end of repetitive sequences. f False positives due to paralogous alignments of reads from regions not well represented in the reference. Alignments for proband include reads with several substitutions relative to the reference sequence within the 41-bp viewing window. This typically occurs when reads from sequences not represented in the reference are mapped to the closest paralog
Orthogonal validation of NGS variants
Whether or not Sanger confirmation should be required for clinically relevant variants remains a matter of debate [80, 81]. In general, the validation rate for NGS variant calls is extremely high—99.965% according to a well-powered study [79]—suggesting that for the vast majority of NGS variants, independent confirmation is unnecessarily redundant. In many cases, a visual manual review of the variant may be enough to determine if it passes muster or warrants orthogonal validation. An interlaboratory study of more than 80,000 clinical specimens demonstrated that a heuristic approach examining fewer than ten criteria (read depth, quality score, observed variant allele sequence, repetitive sequence, etc.) can identify the subset of variants most likely to be false positives and thus requiring orthogonal validation [82].
Identifying de novo mutations
A key advantage of joint calling in trios is the ability to distinguish de novo mutations, which account for a significant proportion of positive diagnoses from clinical genetic testing [11, 83,84,85]. According to recent large-scale trio sequencing studies, the human de novo mutation rate is approximately 1.29 × 10−8 per base pair per generation [86, 87]. Thus, each proband likely harbors ~ 70 de novo mutations genome-wide against a background of ~ 4–5 million inherited variants. In the protein-coding exome, we expect ~ 1 de novo mutation on a background of ~ 50,000 inherited variants. A sequence variant called in the proband is therefore far more likely to be inherited than de novo. Furthermore, even with extremely high variant calling precision (99.9%), there will be 50 false-positive calls for each de novo mutation. Thus, candidate de novo mutations merit careful scrutiny.
In addition to filtering for artifactual calls as described above, de novo mutations should be queried against public databases of genome variation, such as the gnomAD database. Although true de novo mutations can certainly occur at positions of known sequence variants, a candidate de novo with appreciable frequency in the population (i.e., MAF > 0.0001) is far more likely to represent a germline variant. Similarly, manual review in Integrative Genomics Viewer (IGV) should be used to exclude both artifactual calls and variants with supporting evidence in one or both parents (e.g., Fig. 2a).
Copy number and structural variant calling
Copy number variants (CNVs) are a major source of human genetic variation and have been implicated in numerous diseases [88,89,90], such as autism [91], intellectual disability [92], and congenital heart disease [93,94,95]. Although microarray testing is typically ordered prior to panel or exome testing in a clinical setting, NGS-based CNV detection is increasingly incorporated into clinical diagnostic testing and accounts for 3–5% of positive diagnoses. A number of tools exist for identifying CNVs from targeted NGS data, such as cn.MOPS [45], CONTRA [46], CoNVEX [47], ExomeCNV [48], ExomeDepth [49], and XHMM [50]. Most rely on comparisons of sequence depth between a test subject and a comparator to identify significant changes in copy number. Not all CNV calling tools perform well in all situations, and as a rule, the sensitivity for CNV detection using targeted NGS is limited compared to genome sequencing [96].
Paired-end whole-genome sequencing data also enables the detection of structural variants with increasing precision. Popular tools for this application, such as DELLY [51], Lumpy [52], Manta [53], Pindel [54], and SVMerge [55], use two types of information to identify signatures of structural variants. Read pairing information serves to identify segments of the genome in which molecularly linked read pairs map at unexpected distances or orientations. Split read alignments, in which a single sequence read maps to two different regions of the genome, are also incorporated into SV calling. It should be emphasized that while many consider SNV/indel detection with NGS to be routine, SV detection with whole-genome sequencing data is still challenging, as illustrated by the fact that leading tools achieve F-1 values of only ~ 0.80–0.90 in benchmarking experiments. There are at least two principal reasons for this. First, it is widely recognized that a large proportion of structural variation occurs in “difficult” regions of the genome, such as repetitive or tandem-duplicated sequences. Second, the relatively short length of NGS reads (~ 150 bp) and typical fragments (~ 300–500 bp) is often insufficient to resolve complex structural variants and long insertions [97]. For this reason, linked-read and long-read sequencing technologies are increasingly being applied to resolve large SVs and complex sequences [98,99,100], for a recent review, see [101].
Visual review of CNVs and structural variants called by NGS can also, to some extent, be performed in IGV. For SVs in particular, it is useful to view reads as pairs and color them according to insert size, as shown in Fig. 3. Well-supported structural variants are often supported by both discordant read pairs and changes in overall sequence depth, such as the deletions in Fig. 3a and b and the duplication in Fig. 3d. Manual review can also help resolve ambiguous SV breakpoints (Fig. 3c).
Visual review of copy number and structural variants. Each pane is an IGV screenshot of WGS for a proband (top), mother, (middle), and father (bottom). The top track for each sample is a histogram of sequence depth. Reads are viewed as pairs, with discordant pair alignments highlighted in color. a A homozygous ~ 4-kb del that appears heterozygous in the proband, homozygous in the mother, and absent from the father. Note the discordant read pairs suggesting a deletion (red) and visible change in read depth. b Homozygous deletion inherited from two heterozygous parents. c A heterozygous paternally inherited deletion with ambiguous end point by paired-end mapping resolved by visual inspection of read depth. d A maternally inherited tandem duplication. Note the increased read depth in the histogram and the discordant read pairs highlighted in green that span the original sequence and their tandem duplication
Benchmarking germline variant calling pipelines
As described in the previous section, several reference datasets and a “best practice” framework for benchmarking variant calling pipelines are publicly available. At the time of writing, the most recent dataset for sample NA12878 includes ~ 3.04m SNVs and ~ 0.5m small indels, as well as aligned high-depth Illumina sequencing data in BAM format. These resources make it possible to evaluate performance and fine-tune variant calling pipelines to achieve optimal results. For small variants, an F1 score > 0.99 should be achievable by robust variant calling pipelines. High-quality DNA samples for NA12878 can also be ordered from Coriell and sequenced independently to evaluate the performance of a laboratory’s entire pipeline from sample preparation through variant calling.
Benchmarking structural and copy number variant callers tends to be more challenging for two reasons. First, these variants are more challenging to detect with precision using short-read sequencing data. Second, the precise breakpoints for SVs/CNVs are not always well-defined, which makes comparisons across callsets a more complex endeavor. Even so, multiple “gold standard” SV callsets such as GIAB [99], HS1011 [102], and HuRef [103] have been published which employ orthogonal sequencing technologies to define reference callsets comprising thousands of structural variants. When benchmarking with such resources, it is important to recognize that SV calling with short-read data is more error-prone than small variant calling; even the best-performing SV callers only achieve F-1 scores of ~ 0.80–0.90 [103].
Best practices for somatic mutation calling
NGS of tumor specimens is increasingly deployed in oncology to guide diagnosis, prognosis, and personalized care [104]. Although ~ 10% of cancer patients harbor germline predisposition variants, the main purpose of clinical tumor sequencing is often the identification of somatic mutations, copy number alterations, and fusions that may have clinical relevance. A standard pipeline for this is shown in Fig. 1c. It illustrates a paired tumor-normal sequencing strategy, that is, sequencing DNA from a tumor sample and a matched control sample (e.g., blood or skin) from the same patient. Although tumor-only sequencing has been adopted by many laboratories as a cost-effective approach to guide cancer diagnosis, prognosis, and therapy [16, 105,106,107], doing so makes it difficult to distinguish true somatic mutations from constitutional variants [108,109,110]. Thus, the emphasis of this section will be on the “best practice” of sequencing a tumor sample with a matched comparator sample.
Numerous variant callers have been published for this purpose; a list of the most cited callers can be found in Table 2. Widely used somatic mutation callers, such as MuTect2 [40], Strelka2 [42], and VarScan2 [44], consider aligned data from the tumor and normal simultaneously. Several groups have attempted to directly compare the performance of mutation callers for different applications [111,112,113], finding that each has strengths and weaknesses. Because no somatic caller has emerged which offers superior performance in all scenarios, an ensemble approach that combines the results of two or more complementary callers may offer the best balance of sensitivity and specificity [73, 114].
Several aspects of clinical tumor sequencing can make the detection of somatic mutations more challenging. Tumor purity—the proportion of cells in a sample that are cancerous—governs the representation of somatic mutations in a sequenced sample, but pathology estimates of purity based on light microscopy are notoriously inaccurate [115,116,117]. Somatic mutations present at low frequency due to low tumor cellularity and/or subclonal mutation architectures can be challenging to detect, even with high-depth sequencing data. Although many somatic mutation callers such as VarScan2 can be configured for the detection of variants at low frequencies, doing so often reduces the overall false-positive rate. The type of specimen obtained for sequencing also influences mutation calling. Formalin-fixed, paraffin-embedded (FFPE) samples, which are preferred for histopathological diagnosis, often harbor thousands of artifacts arising from chemical DNA damage [118,119,120]. These challenges call for a robust somatic mutation detection pipeline that performs well across many types of clinical tumor samples.
Filtering somatic variant calls
Similar to germline SNVs/indels, candidate somatic variants should be filtered to remove common alignment artifacts such as those illustrated in Fig. 2. In addition, the availability of a matched normal sample enables a direct comparison of data characteristics at the site of a candidate somatic variant call to help distinguish true variants from false positives. For example, reads supporting high-quality mutation calls should exhibit similar position and strandedness as reads supporting the wild-type allele. Other metrics, such as the difference in average mapping quality or trimmed read length, help uncover false positives due to alignment artifacts. Mismatch quality sum (MMQS) difference, computed as the average sum of base qualities for non-reference base calls in variant-supporting reads, is a powerful metric for identifying false positives associated with paralogous alignments [121].
Filtering with population databases
Population variant filtering is a powerful strategy for identifying and removing likely germline variants from somatic mutation callsets but should be done with caution. Simply removing all variants in dbSNP [122] is an appealing but hazardous strategy, since that database contains a number of recurrent mutations from human tumors—such as p.(H1047R) in PIK3CA (rs121913279) and p.(R132H) in IDH1 (rs121913500)—as well as several mutations from the COSMIC somatic mutation database [109]. There is a similar risk for applying a broad filter based on all variants in the gnomAD database [123], in which the presence of apparent somatic loss-of-function variants in hematological malignancy genes like ASXL1 has been documented [124]. Allele frequency information can be used to safeguard against the inadvertent filtering of true somatic variants that are present in such databases. Requiring a minimum minor allele frequency > 0.0001 in the gnomAD or TopMed database is recommended to select variants for filtering somatic mutation callsets.
Some groups have also found value in using an internal “panel of normals” to identify and remove recurrent sequencing artifacts [38]. In this approach, sequencing data from a set of normal DNA specimens (typically ~ 50) are compiled into a reference panel against which candidate somatic variants from tumors can be quickly filtered to remove variant calls associated with germline variants or sequencing artifacts. This approach is advantageous because it identifies artifacts that may be specific to a laboratory’s sequencing protocols or downstream analysis pipelines.
High-confidence somatic SNV/indel calls
In summary, high-confidence somatic SNV/indel calls should be identified by multiple somatic mutation calling tools at positions with sufficient sequencing coverage (> 10× in both tumor and normal tissue). Variant alleles should be supported by reads on both strands with no apparent bias in read position, base quality, or mapping quality. High-quality SNVs/indels should also be absent from public databases and an internal laboratory panel of normal (if available), or else present at very low frequencies (MAF < 0.001). Finally, candidate SNV/indel calls should be reviewed by visualization of the tumor and normal sequencing alignments with a tool such as IGV.
Calling somatic copy number and structural variants
Many of the tools developed for germline CNV/SV calling have been adapted for cancer genomics [125], and still, others have been developed for the critical task of identifying fusions from RNA-seq data [126]. Somatic copy number alteration (SCNA) detection is arguably the easier of the two tasks, since a matched normal sample is often alive to use as a comparator. Further, deep sequencing data allow for precise determination of variant allele frequencies, the skewing of which can often be observed to support candidate variants. Similar to somatic mutation calling, combining the results of at least two tools, such as VarScan 2 (less conservative) and GATK (more conservative), may provide the optimal strategy for calling somatic CNAs. Further, incorporation of tumor variant allele frequency (VAF) information can help generate supporting evidence for somatic structural variants, since changes in copy number tend to skew allele frequencies of heterozygous variants (Fig. 4). Similar to somatic SNV/indel calling, somatic SV/CNA calls may be filtered against a panel of normals to remove calls in regions of highly variable copy number and recurrent artifactual SVs.
Detecting somatic rearrangements in cancer using NGS. Shown is whole-genome sequencing data for chromosome 1 for a tumor-normal pair. Top: Log2 values indicate copy number changes in the tumor relative to the normal. Bottom: copy gains and losses skew tumor allele frequencies for heterozygous variants, with loss of heterozygosity (red) apparent in regions of heterozygous deletions
Benchmarking somatic calling pipelines
Benchmarking somatic mutation callers requires a reference “truth set” of real somatic mutations. Such datasets have been generated by synthetic mixing experiments (for example, of NA12878 with another well-characterized sample at specifically defined proportions). Of note, though numerous comparisons of somatic mutation callers have been published, the findings are inconsistent [127]. One reason for this is that the researchers conducting those studies often apply variant callers with default parameter settings or neglect to perform critical downstream filtering. To address this issue, the DREAM ICGC-TCGA Somatic Mutation calling challenge invited teams, including several developers of somatic mutation calling tools, to benchmark their pipelines on a common dataset. The organizers employed a robust simulation framework to introduce synthetic somatic alterations (i.e., a truth set) into real WGS data for three tumors upon which each team’s submissions were evaluated. The simulated datasets and truth sets from these challenges are freely available and offer a well-vetted benchmarking resource for somatic SNV, indel, and structural variant calling pipelines [128].
Conclusions and future directions
Variant calling in NGS data, much like NGS technologies themselves, has evolved considerably over the past decade and remains an active area of research. Robust pipelines for NGS analysis include steps for optimized alignment and pre-processing, variant calling, filtering of false positives, and visual manual review. While some of these procedures, such as read alignment and SNV/indel detection, can be suitably performed with a single software package, others, such as CNV/SV calling and somatic mutation detection, benefit from incorporating multiple independent tools. Benchmarking resources for both germline and somatic variants provide an opportunity to evaluate and optimize the performance of variant calling. Although some classes of variants—such as de novo mutations in germline studies and low-frequency somatic mutations in cancer patients—likely require validation on an orthogonal platform, the burden of additional confirmatory testing is likely to decrease as technologies continue to improve. However, the observation that even state-of-the-art SV callers only achieve F-scores of ~ 0.80–0.90 in gold standard datasets suggests that emerging long-read sequencing technologies may ultimately be required to accurately call large and/or complex structural variants. Nevertheless, the general principles discussed in this review—rigorous pre-processing of sequencing data, implementation of multiple variant calling approaches, and systematic filtering to remove artifacts—will remain relevant guidance for clinical variant calling in years to come.
Availability of data and materials
Not applicable
Abbreviations
- BAM:
-
Binary alignment/map
- CNV:
-
Copy number variant
- NGS:
-
Next-generation sequencing
- SNV:
-
Single nucleotide variant
- VCF:
-
Variant call format
References
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70.
Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7.
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15.
Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9.
Cancer Genome Atlas Research N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73.
Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74.
Cancer Genome Atlas Research N. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–20.
Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, et al. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet A. 2012;158A(7):1523–5.
Consortium UK, Walter K, Min JL, Huang J, Crooks L, Memari Y, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90.
Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Tippin Davis B, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2015;17(7):578–86.
Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18(7):696–704.
Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, Weiss ME, Koster J, Marais A, et al. Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet. 2017;25(2):176–82.
Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369(16):1502–11.
Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2016;2(5):616–24.
Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–31.
Wheler J, Lee JJ, Kurzrock R. Unique molecular landscapes in cancer: implications for individualized, curated drug combinations. Cancer Res. 2014;74(24):7181–4.
Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135(4):441–50.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8.
Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, Consortium WGS, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46(8):912–8.
de Ligt J, Boone PM, Pfundt R, Vissers LE, Richmond T, Geoghegan J, et al. Detection of clinically relevant copy number variants with whole-exome sequencing. Hum Mutat. 2013;34(10):1439–48.
Pagnamenta AT, Lise S, Harrison V, Stewart H, Jayawant S, Quaghebeur G, et al. Exome sequencing can detect pathogenic mosaic mutations present at low allele frequencies. J Hum Genet. 2012;57(1):70–2.
Qin L, Wang J, Tian X, Yu H, Truong C, Mitchell JJ, et al. Detection and quantification of mosaic mutations in disease genes by next-generation sequencing. J Mol Diagn. 2016;18(3):446–53.
Shin HT, Choi YL, Yun JW, Kim NKD, Kim SY, Jeon HJ, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017;8(1):1377.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9.
Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100.
Institute B. Picard Tools. http://broadinstitute.github.io/picard. Accessed 1 Oct 2019.
Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032–4.
Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30(17):2503–5.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2.
Okonechnikov K, Conesa A, Garcia-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32(2):292–4.
Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–73.
Jun G, Flickinger M, Hetrick KN, Romm JM, Doheny KF, Abecasis GR, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91(5):839–48.
Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012;1207.3907. https://arxiv.org/abs/1207.3907v2.
Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–93.
Gerstung M, Beisel C, Rechsteiner M, Wild P, Schraml P, Moch H, et al. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat Commun. 2012;3:811.
Fan Y, Xi L, Hughes DS, Zhang J, Zhang J, Futreal PA, et al. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol. 2016;17(1):178.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–9.
Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28(3):311–7.
Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–7.
Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44(11):e108.
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76.
Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, et al. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40(9):e69.
Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28(10):1307–13.
Amarasinghe KC, Li J, Halgamuge SK. CoNVEX: copy number variation estimation in exome sequencing data using HMM. BMC Bioinformatics. 2013;14(Suppl 2):S2.
Sathirapongsasuti JF, Lee H, Horst BA, Brunner G, Cochran AJ, Binder S, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics. 2011;27(19):2648–54.
Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28(21):2747–54.
Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91(4):597–607.
Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28(18):i333–i9.
Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15(6):R84.
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220–2.
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25(21):2865–71.
Wong K, Keane TM, Stalker J, Adams DJ. Enhanced structural variant and breakpoint detection using SVMerge by integration of multiple detection methods and local assembly. Genome Biol. 2010;11(12):R128.
Nicorici D, Şatalan M, Edgren H, Kangaspeska S, Murumägi A, Kallioniemi O, et al. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data. BioRxiv. 2014. https://doi.org/10.1101/011650.
Ge H, Liu K, Juan T, Fang F, Newman M, Hoeck W. FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27(14):1922–8.
Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38(18):e178.
Jia W, Qiu K, He M, Song P, Zhou Q, Zhou F, et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 2013;14(2):R12.
Haas BJ, Dobin A, Stransky N, Li B, Yang X, Tickle T, et al. STAR-Fusion: fast and accurate fusion transcript detection from RNA-Seq. BioRxiv. 2017. https://doi.org/10.1101/120295.
Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12(8):R72.
Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28(4):464–9.
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6.
Shigemizu D, Momozawa Y, Abe T, Morizono T, Boroevich KA, Takata S, et al. Performance comparison of four commercial human whole-exome capture platforms. Sci Rep. 2015;5:12742.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 0 1–33.
Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30(20):2843–51.
Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32(3):246–51.
Eberle MA, Fritzilas E, Krusche P, Kallberg M, Moore BL, Bekritsky MA, et al. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. 2017;27(1):157–64.
Zook JM, McDaniel J, Olson ND, Wagner J, Parikh H, Heaton H, et al. An open resource for accurately benchmarking small variant and reference calls. Nat Biotechnol. 2019;37(5):561–6.
Krusche P, Trigg L, Boutros PC, Mason CE, De La Vega FM, Moore BL, et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat Biotechnol. 2019;37(5):555–60.
Li H, Bloom JM, Farjoun Y, Fleharty M, Gauthier L, Neale B, et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat Methods. 2018;15(8):595–7.
Toptas BC, Rakocevic G, Komar P, Kural D. Comparing complex variants in family trios. Bioinformatics. 2018;34(24):4241–7.
Callari M, Sammut SJ, De Mattos-Arruda L, Bruna A, Rueda OM, Chin SF, et al. Intersect-then-combine approach: improving the performance of somatic variant calling in whole exome sequencing data using multiple aligners and callers. Genome Med. 2017;9(1):35.
Highnam G, Wang JJ, Kusler D, Zook J, Vijayan V, Leibovich N, et al. An analytical framework for optimizing variant discovery from personal genomes. Nat Commun. 2015;6:6275.
Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;5:17875.
Sandmann S, de Graaf AO, Karimi M, van der Reijden BA, Hellstrom-Lindberg E, Jansen JH, et al. Evaluating variant calling tools for non-matched next-generation sequencing data. Sci Rep. 2017;7:43169.
Yohe S, Hauge A, Bunjer K, Kemmer T, Bower M, Schomaker M, et al. Clinical validation of targeted next-generation sequencing for inherited disorders. Arch Pathol Lab Med. 2015;139(2):204–10.
Baudhuin LM, Lagerstedt SA, Klee EW, Fadra N, Oglesbee D, Ferber MJ. Confirming variants in next-generation sequencing panel testing by sanger sequencing. J Mol Diagn. 2015;17(4):456–61.
Beck TF, Mullikin JC, Program NCS, Biesecker LG. Systematic evaluation of sanger validation of next-generation sequencing variants. Clin Chem. 2016;62(4):647–54.
Mu W, Lu HM, Chen J, Li S, Elliott AM. Sanger confirmation is required to achieve optimal sensitivity and specificity in next-generation sequencing panel testing. J Mol Diagn. 2016;18(6):923–32.
Strom SP, Lee H, Das K, Vilain E, Nelson SF, Grody WW, et al. Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory. Genet Med. 2014;16(7):510–5.
Lincoln SE, Truty R, Lin CF, Zook JM, Paul J, Ramey VH, et al. A rigorous interlaboratory examination of the need to confirm next-generation sequencing-detected variants with an orthogonal method in clinical genetic testing. J Mol Diagn. 2019;21(2):318–29.
Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312(18):1880–7.
Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376(1):21–31.
Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17(10):774–81.
Besenbacher S, Liu S, Izarzugaza JM, Grove J, Belling K, Bork-Jensen J, et al. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat Commun. 2015;6:5969.
Jonsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549(7673):519–22.
Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330(6004):641–6.
Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84(2):148–61.
Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009;93(1):22–6.
Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5(6):e1000536.
Whibley AC, Plagnol V, Tarpey PS, Abidi F, Fullston T, Choma MK, et al. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am J Hum Genet. 2010;87(2):173–88.
Soemedi R, Wilson IJ, Bentham J, Darlay R, Topf A, Zelenika D, et al. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet. 2012;91(3):489–501.
Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, et al. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci U S A. 2011;108(7):2915–20.
Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41(8):931–5.
Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu X, Guo H, et al. An evaluation of copy number variation detection tools from whole-exome sequencing data. Hum Mutat. 2014;35(7):899–907.
Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363–76.
Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10(1):1784.
Zook JM, Hansen NF, Olson ND, Chapman L, Mullikin JC, Xiao C, et al. A robust benchmark for detection of germline large deletions and insertions. Nat Biotechnol. 2020; https://doi.org/10.1038/s41587-020-0538-8.
Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, et al. Characterizing the major structural variant alleles of the human genome. Cell. 2019;176(3):663–75 e19.
Ho SS, Urban AE, Mills RE. Structural variation in the sequencing era. Nat Rev Genet. 2020;21(3):171–89.
English AC, Salerno WJ, Hampton OA, Gonzaga-Jauregui C, Ambreth S, Ritter DI, et al. Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics. 2015;16:286.
Mu JC, Tootoonchi Afshar P, Mohiyuddin M, Chen X, Li J, Bani Asadi N, et al. Leveraging long read sequencing from a single individual to provide a comprehensive resource for benchmarking variant calling methods. Sci Rep. 2015;5:14493.
Giardina T, Robinson C, Grieu-Iacopetta F, Millward M, Iacopetta B, Spagnolo D, et al. Implementation of next generation sequencing technology for somatic mutation detection in routine laboratory practice. Pathology. 2018;50(4):389–401.
Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–64.
Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16(1):56–67.
Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93.
Garofalo A, Sholl L, Reardon B, Taylor-Weiner A, Amin-Mansour A, Miao D, et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med. 2016;8(1):79.
Hiltemann S, Jenster G, Trapman J, van der Spek P, Stubbs A. Discriminating somatic and germline mutations in tumor DNA samples without matching normals. Genome Res. 2015;25(9):1382–90.
Sukhai MA, Misyura M, Thomas M, Garg S, Zhang T, Stickle N, et al. Somatic tumor variant filtration strategies to optimize tumor-only molecular profiling using targeted next-generation sequencing panels. J Mol Diagn. 2019;21(2):261–73.
Wang Q, Jia P, Li F, Chen H, Ji H, Hucks D, et al. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med. 2013;5(10):91.
Xu H, DiCarlo J, Satya RV, Peng Q, Wang Y. Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genomics. 2014;15:244.
Kroigard AB, Thomassen M, Laenkholm AV, Kruse TA, Larsen MJ. Evaluation of nine somatic variant callers for detection of somatic mutations in exome and targeted deep sequencing data. PLoS One. 2016;11(3):e0151664.
Fang LT, Afshar PT, Chhibber A, Mohiyuddin M, Fan Y, Mu JC, et al. An ensemble approach to accurately detect somatic mutations using SomaticSeq. Genome Biol. 2015;16:197.
Viray H, Li K, Long TA, Vasalos P, Bridge JA, Jennings LJ, et al. A prospective, multi-institutional diagnostic trial to determine pathologist accuracy in estimation of percentage of malignant cells. Arch Pathol Lab Med. 2013;137(11):1545–9.
Smits AJ, Kummer JA, de Bruin PC, Bol M, van den Tweel JG, Seldenrijk KA, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol. 2014;27(2):168–74.
Dudley J, Tseng LH, Rooper L, Harris M, Haley L, Chen G, et al. Challenges posed to pathologists in the detection of KRAS mutations in colorectal cancers. Arch Pathol Lab Med. 2015;139(2):211–8.
Bass BP, Engel KB, Greytak SR, Moore HM. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen? Arch Pathol Lab Med. 2014;138(11):1520–30.
Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61(1):64–71.
Oh E, Choi YL, Kwon MJ, Kim RN, Kim YJ, Song JY, et al. Comparison of accuracy of whole-exome sequencing with formalin-fixed paraffin-embedded and fresh frozen tissue samples. PLoS One. 2015;10(12):e0144162.
Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for germline variant calling and somatic mutation detection. Curr Protoc Bioinformatics. 2013;44:15 4 1–7.
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11.
MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335(6070):823–8.
Carlston CM, O’Donnell-Luria AH, Underhill HR, Cummings BB, Weisburd B, Minikel EV, et al. Pathogenic ASXL1 somatic variants in reference databases complicate germline variant interpretation for Bohring-Opitz syndrome. Hum Mutat. 2017;38(5):517–23.
Alkodsi A, Louhimo R, Hautaniemi S. Comparative analysis of methods for identifying somatic copy number alterations from deep sequencing data. Brief Bioinform. 2015;16(2):242–54.
Liu S, Tsai WH, Ding Y, Chen R, Fang Z, Huo Z, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016;44(5):e47.
Xu C. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput Struct Biotechnol J. 2018;16:15–24.
Lee AY, Ewing AD, Ellrott K, Hu Y, Houlahan KE, Bare JC, et al. Combining accurate tumor genome simulation with crowdsourcing to benchmark somatic structural variant detection. Genome Biol. 2018;19(1):188.
Acknowledgements
The author wishes to thank Richard K. Wilson, Elaine R. Mardis, Catherine E. Cottrell, Peter White, and other members of the Steve and Cindy Institute for Genomic Medicine at Nationwide Children’s Hospital.
Funding
This work was generously supported by the Pediatric Innovation Fund, under the auspices of the Research Institute at Nationwide Children’s Hospital.
Author information
Authors and Affiliations
Contributions
DCK wrote the manuscript in its entirety. The author read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The author is a co-inventor of VarScan 2 and thus receives a portion of licensing royalties from several commercial licensees.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Koboldt, D.C. Best practices for variant calling in clinical sequencing. Genome Med 12, 91 (2020). https://doi.org/10.1186/s13073-020-00791-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13073-020-00791-w